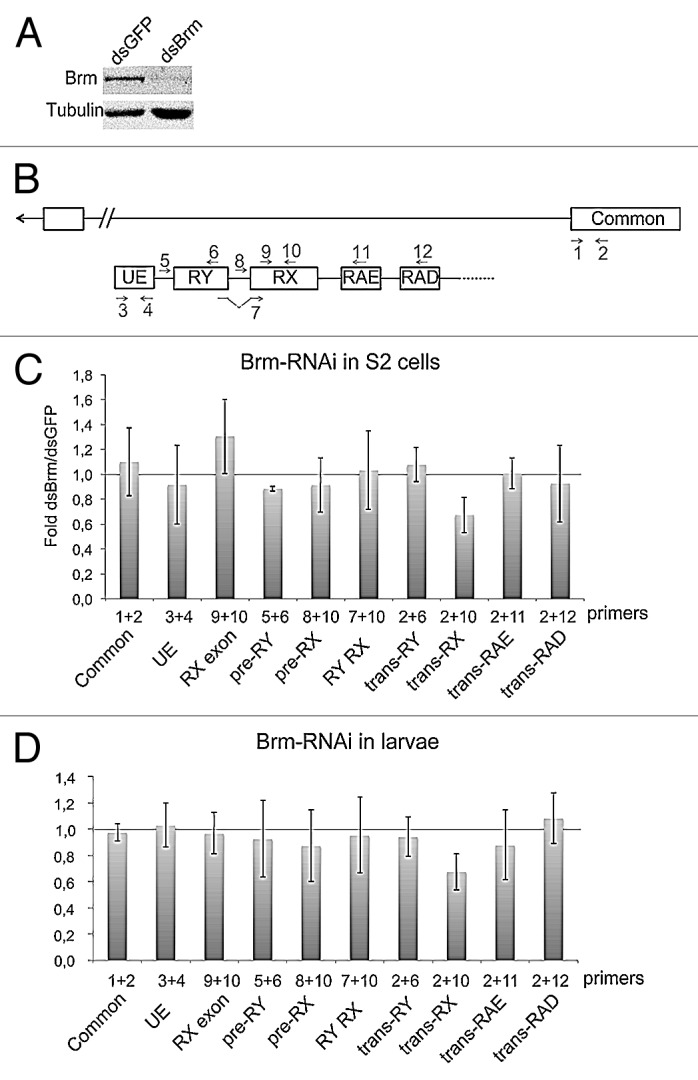
Figure 5. Depletion of Brm by RNAi reduces the abundance of trans-spliced RX transcripts. (A) RNAi was used to deplete BRM in S2 cells. Cells were treated in parallel with dsGFP as a control. The effect of BRM depletion was analyzed by western blotting after 48 h of dsRNA treatment. Tubulin was used as loading control. (B) Scheme showing the organization of the mod(mdg4) locus and the positions of the primer-pairs used in the PCR experiments that are presented in this figure. (C) The relative abundances of sense and anti-sense transcripts were quantified by RT-qPCR and calculated relative to Act5C. The ratios between dsBrm-treated cells and dsGFP-treated cells are presented in the histogram. The bars represent averages from five independent experiments. The error bars represent standard deviations. The trans-RX transcript was significantly reduced (P = 0.014, comparison of transcript abundances calculated relative to Act5C mRNA using a two-tailed, paired Student’s t test). The abundance of the other analyzed transcripts was not significantly changed (P > 0.05). (D) The effect of BRM depletion on the mod(mdg4) trans-splicing in larvae. hs-GAL4 virgin females were crossed with UAS-BrmRNAi males, and early third instar larvae from the cross were heat shocked for 2 h at 37 °C. Control experiments were performed in parallel by crossing hs-GAL4 virgin females with wild type males W1118. Total RNA was purified and the abundances of selected transcripts were analyzed by RT-qPCR, as in (C). The bars represent averages from three independent experiments. The trans-RX transcript was significantly reduced (P = 0.017, comparison of transcript abundances calculated relative to the common exon 4 using a two-tailed, Student’s t test).
