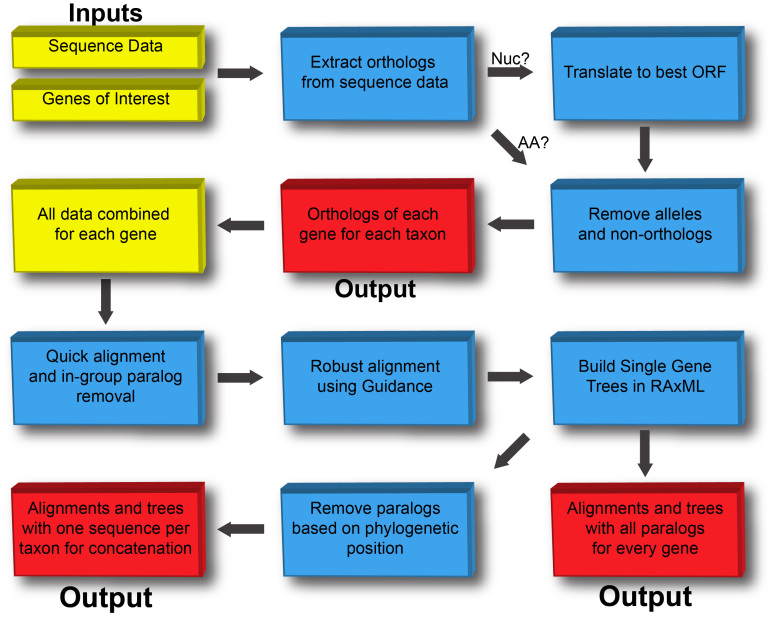
Flowchart showing major steps in the pipeline.
First, the scripts relating to Taxon Objects output orthologs for each gene of interest by starting with sequence data from target taxa and fasta files of orthogs from OrthoMCL. The orthologs are then combined into Gene Objects and a series of refinement steps are performed including removal of ingroup paralogs, alignment with Guidance (Penn et al., 2010) and generation of single gene trees. The outputs are alignments and trees with 1) all paralogs and 2) paralogs removed in preparation for concatenation.