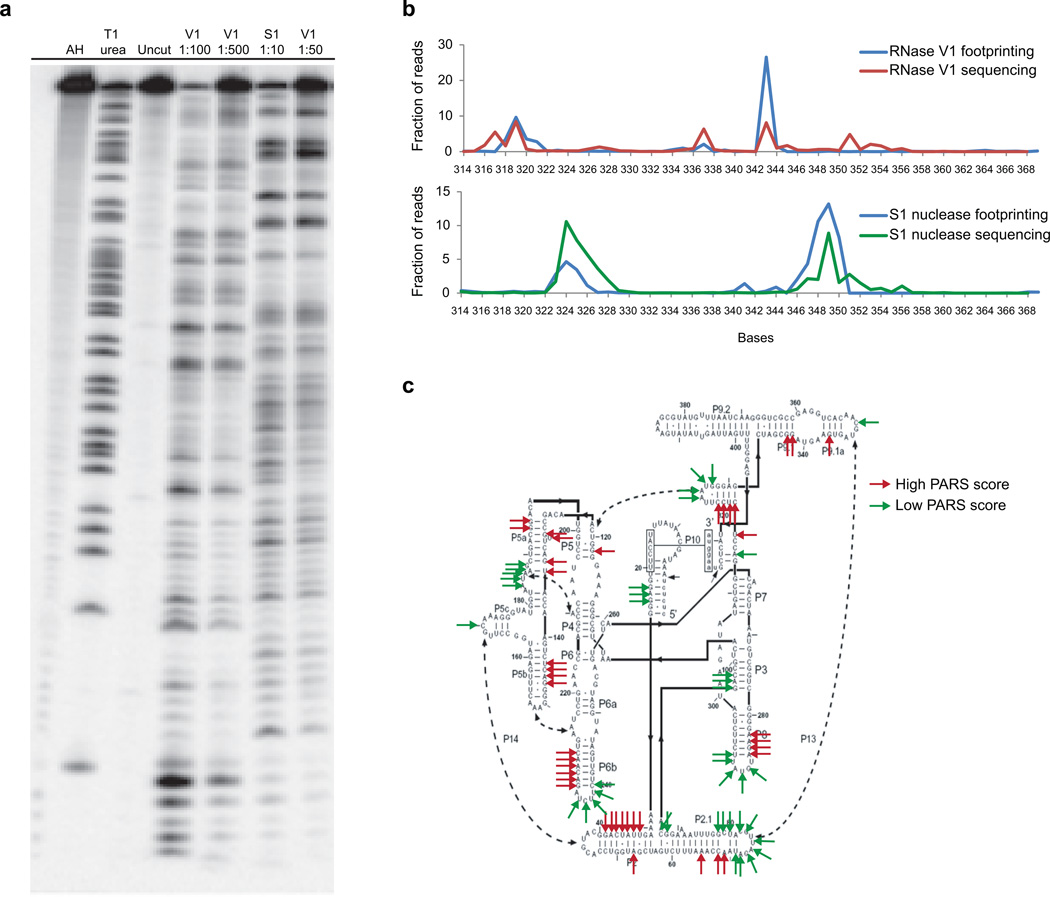
Extended Data Figure 1. PARS data accurately maps to known structures.
a, RNase V1 and S1 nucleases were titrated to single hit kinetics in structure probing. Gel analysis of structure probing of yeast RNA in the presence of 1μg of total human RNA using different dilutions of RNase V1 (lanes 4, 5), and S1 nuclease (lanes 6,7), cleaved at 37°C for 15min. Additionally, RNase T1 ladder (lane 2), alkaline hydrolysis (lane 1), and no nuclease treatment (lane3) are shown. Dilution of V1 nuclease by 1:500 and S1 nuclease by 1:50 results in mostly intact RNA. b, PARS signal obtained for the P9-9.2 domain of Tetrahymena ribozyme using the double strand enzyme RNase V1 (red line) or the single strand enzyme S1 nuclease (green line) accurately matches the signals obtained by tranditional footprinting (blue lines). c, Top 10 percentile of PARS score (double stranded, red arrows) and bottom 10 percentile of PARS score (single stranded, green arrows) were mapped to the secondary structure of the Tetrahymena ribozyme.