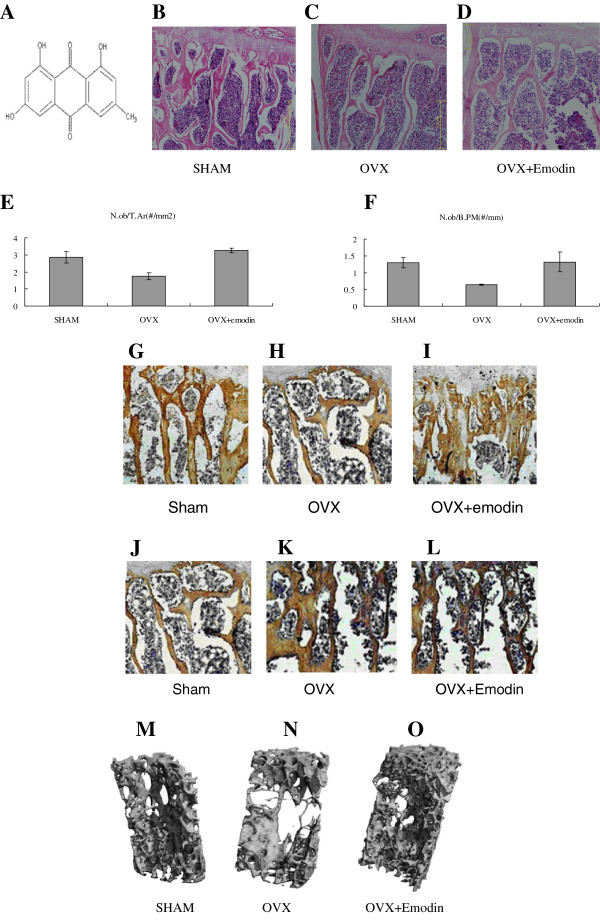
Figure 1.
The structure of emodin. (A) Molecular formula: C15H10O5, molecular weight: 270.23. HE Staining showed more osteoblasts attaching to the surface of bone trabeculae of emodin-OVX group (C) and SHAM group (B) than OVX group (D). Osteoblast activity scores from HE staining slices: N.ob/T.Ar (osteoblast number per trabecular area). (E) and N.ob/B.PM (osteoblast number per bone perimeter) (F) rose up in OVX + emodin group. The columns represent the mean ± SE of three independent experiments. *: P < 0.05 vs. OVX. Immunohistochemical assessments of the protein of Runx2 (G, H, I) and PPARγ (J, K, L). Immunohistochemical analysis showed more increase in Runx2 protein distribution in the OVX + emodin groups than OVX group. Immunohistochemical analysis showed more increase in PPARγprotein distribution in the OVX + emodin groups. Representative 3-D images of lumbar vertebral body of sham-operated mice (M:SHAM), ovariectomy mice (N:OVX) and emodin-treated OVX mice (O:OVX + emodin) by micro-CT. Bone loss and trabecular bone deterioration are evident in the OVX group and the progression of bone loss and bone deterioration is partially rescued by emodin treatment.