Table 1.
List of proteins with different abundance in cancer cell lines
|
Protein name |
Ascension number (Swissprot)/matched 2D coordinates |
MW/pI |
Peptides (% cov) |
Relative change vs normal |
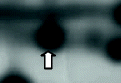
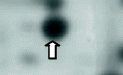
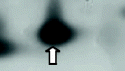
Zoomed in gel image showing representative spots |
|||
|---|---|---|---|---|---|---|---|---|
|
Abbreviation |
(World-2DPAGE portal) |
|||||||
| Theory | Experimental | Normal | Cancer | |||||
| Structural | ||||||||
| 20 |
Stathmin (Phosphoprotein p19) |
P16949 /SPOT 2D-001YH8 |
17.3/5.76 |
>15.0/5.7 |
4 (18%) |
+1.6 |
 |
 |
| STMN1 | ||||||||
| 1 |
Tropomyosin alpha-3 chain (Tropomyosin-3) |
P06753 / |
32.8/4.68 |
>30.0/4.6 |
6 (13%) |
-1.5 |
 |
 |
| SPOT 2D-001EM1 | ||||||||
| SPOT 2D-001YBH | ||||||||
| TPM3 |
SPOT 2D-001JNO |
|||||||
| 4 |
Myosin regulatory light chain-2 |
P19105 / |
19.8/4.67 |
>15.0/4.60 |
5 (17%) |
-1.9 |
 |
 |
| SPOT 2D-001JUH | ||||||||
| ML12A |
SPOT 2D-001JUS |
|||||||
| Enzymes | ||||||||
| 6 |
Lactoylglutathione lyase (Glyoxalase I). (EC 4.4.1.5) |
Q04760 / |
20.8/5.12 |
>20.0/5.00 |
10 (22%) |
+7.7 |
 |
 |
| LGUL |
n/a |
|||||||
| 24 |
Triosephosphate isomerase (EC 5.3.1.1) |
P60174 / |
26.7/6.45 |
>25.0/6.5 |
5 (28%) |
+2.5 |
 |
 |
| TPIS |
SPOT 2D-0003MP |
|||||||
| 18 |
Nucleoside diphosphate kinase A (EC 2.7.4.6) |
P15531 / |
17.1/5.83 |
>15.0/5.8 |
8 (31%) |
+1.5 |
 |
 |
| NDKA |
n/a |
|||||||
| 21 |
Ubiquitin-conjugating enzyme E2 N (EC 6.3.2.19) |
P61088 / |
17.1/6.13 |
>15.0/5.9 |
6 (23%) |
+2.6 |
 |
 |
| UBE2N |
n/a |
|||||||
| 16 |
Gamma-glutamylcyclotransferase (EC 2.3.2.4) |
O75223 / |
21.0/5.07 |
>20.0/5.0 |
3 (27%) |
+2.5 |
 |
 |
| GGCT |
n/a |
|||||||
| 14 |
Peroxiredoxin-4 (EC 1.11.1.15) |
Q13162 / |
30.5/5.86 |
>30.0/5.8 |
6 (16%) |
+2.2 |
 |
 |
| PRDX4 |
n/a |
|||||||
| 19 |
Superoxide dismutase [Cu-Zn] (EC 1.15.1.1) |
P00441 / |
15.9/5.70 |
>15.0/5.70 |
5 (24%) |
+1.4 |
 |
 |
| SPOT 2D-001F1V | ||||||||
| SODC |
SPOT 2D-000ZXR |
|||||||
| 17 |
Glutathione S-transferase P (GST class-pi). (EC 2.5.1.18) |
P09211 / |
23.3/5.43 |
>25/5.6 |
8 (22%) |
+1.4 |
 |
 |
| SPOT 2D-001EUM | ||||||||
| SPOT 2D-0003Q9 | ||||||||
| GSTP1 |
SPOT 2D-00085X |
|||||||
| 11 |
Proteasome subunit beta type 4 precursor. (EC 3.4.25.1) |
P28070 / |
29.2/5.72 |
29.0/5.7 |
6 (15%) |
+1.6 |
 |
 |
| SPOT 2D-000851 | ||||||||
| PSB4 |
SPOT 2D-000D4M |
|||||||
| Regulatory proteins | ||||||||
| 3 |
14-3-3 protein beta/alpha |
P31946 / |
28.0/4.70 |
>25.0/4.50 |
6 (4%) |
+1.8 |
 |
 |
| 1433B |
n/a |
|||||||
| 12 |
Prohibitin |
P35232 / |
29.8/5.57 |
>25.0/5.60 |
5 (12%) |
+1.6 |
 |
 |
| PHB |
SPOT 2D-001EOL |
|||||||
| 13 |
Proteasome activator complex subunit 1 |
Q06323 / |
28.7/5.78 |
>25.0/5.70 |
2 (4%) |
-1.7 |
 |
 |
| PSME1 |
SPOT 2D-001YC7 |
|||||||
| 16 |
Proteasome activator complex subunit 2 |
Q9UL46 / |
27.3/5.44 |
>30.0/5.4 |
4 (11%) |
-2.3 |
 |
 |
| PSME2 |
n/a |
|||||||
| 2 |
14-3-3 protein sigma (Stratifin) |
P31947 / |
27.8/4.68 |
>25.0/4.50 |
9 (24%) |
-2.0 |
 |
 |
| 1433S |
SPOT 2D-001EP6 |
|||||||
| 22 |
Stress-induced-phosphoprotein 1 |
P31948 / |
62.6/6.4 |
>55.0/6.50 |
11 (12%) |
+2.1 |
 |
 |
| STIP1 |
n/a |
|||||||
| 10 |
Interleukin-1 receptor antagonist protein precursor |
P18510 / |
20.0/5.83 |
>20.0/5.3 |
2 (8%) |
+1.3 |
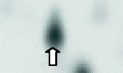 |
 |
| IL1RA |
n/a |
|||||||
| 5 |
Rho GDP-dissociation inhibitor 1 |
P52565 / |
23.2/5.02 |
30.0/5.0 |
4 (14%) |
+2.6 |
 |
 |
| GDIR |
n/a |
|||||||
| 9 |
Ran-specific GTPase-activating protein |
P43487 / |
23.3/5.19 |
30/5.2 |
5 (18%) |
+1.8 |
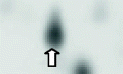 |
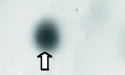 |
| RANG |
n/a |
|||||||
| Chaperones | ||||||||
| 15 |
Heat-shock protein beta-1 |
P04792 / |
22.8/6.00 |
>20.0/5.8 |
11 (32%) |
+3.1 |
 |
 |
| SPOT 2D-001JQI | ||||||||
| HSPB1 |
SPOT 2D-001JQP |
|||||||
| 23 |
T-complex protein 1 subunit zeta-2 |
Q92526 / |
57.7/6.63 |
>55.0/6.8 |
2 (2%) |
-1.6 |
 |
 |
| TCPW |
n/a |
|||||||
| Others | ||||||||
| 8 |
Chloride intracellular channel protein 1 (membrane associated) |
O00299 / |
26.9/5.09 |
>30.0/5.1 |
14 (46%) |
-1.7 |
 |
 |
| CLIC1 | SPOT 2D-001YBL | |||||||
The proteins were grouped into five functional categories – structural proteins, enzymes, regulatory proteins, chaperones and others. The accession number (swiss-prot) and matched 2D coordinates, molecular weight and pI (theoretical and experimental), number of matched peptides, percentage coverage, relative change vs normal and zoomed in gel image showing representative spots are shown.
