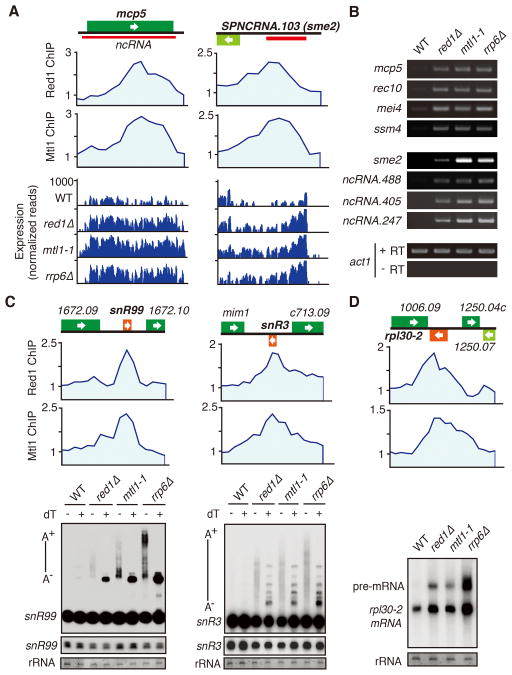
Figure 3. MTREC Regulates Gene Expression and Targets Noncoding RNAs and premRNAs Degraded by the Exosome.
(A) ChIP-chip analysis of Red1-MYC and Mtl1-MYC distribution at indicated loci (top). RNA-Seq analysis of Red1 and Mtl1 bound genes in red1Δ, mtl1-1, and rrp6Δ strains (bottom). (B) RT-PCR analysis of Red1 and Mtl1 bound loci in indicated strains. The transcript level of act1 was used as a control. +RT and −RT indicate presence or absence of reverse transcriptase. (C) ChIP-chip analysis of Red1-MYC and Mtl1-MYC distribution at snoRNAs (top). Northern analysis of snR99 and snR3 processing in mutant strains (bottom). −/+ dT denotes RNase H treatment in the absence or presence of oligo dT. A− /A+ indicates non-polyadenylated and polyadenylated species. Lighter exposures of mature snR99 and snR3 are shown below. rRNA was used as a loading control. (D) Red1-MYC and Mtl1-MYC distribution at rpl30-2 (top). Northern blot analysis of rpl30-2 pre-mRNA processing in the indicated strains (bottom). See also Table S2.