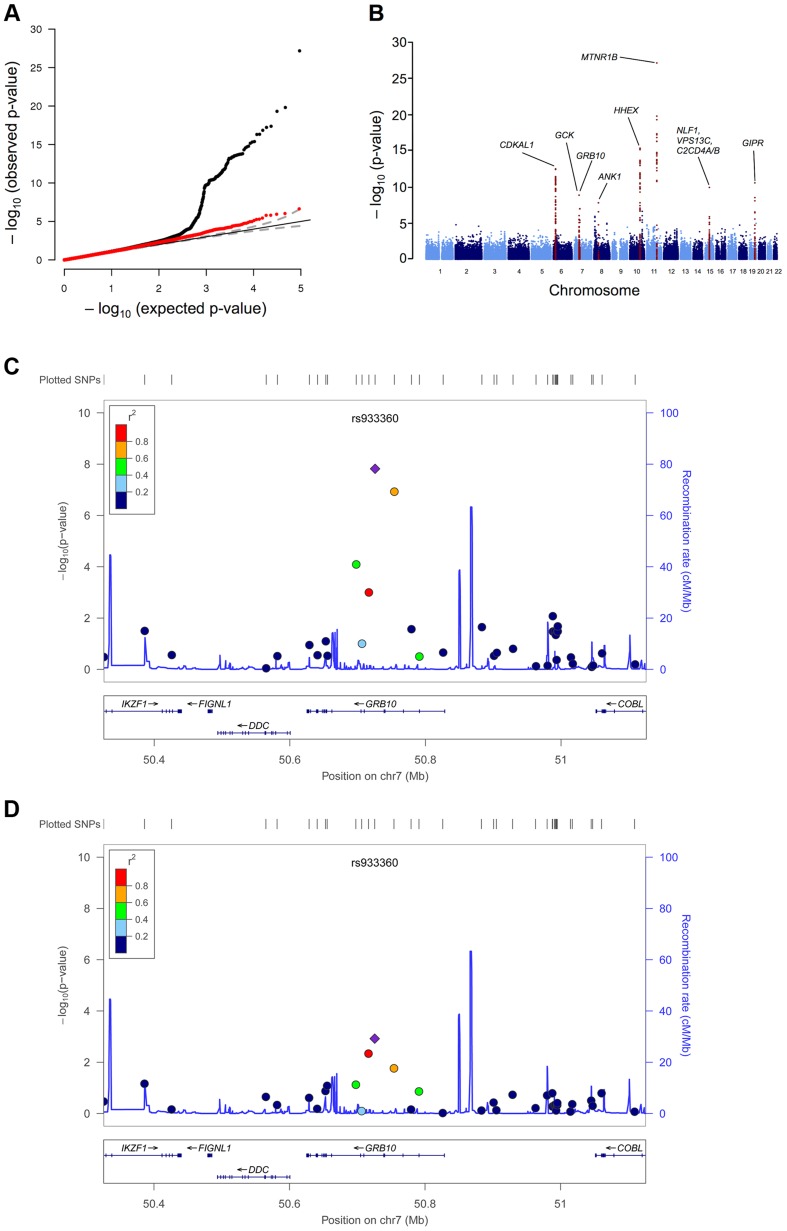
Figure 1. GWAS plots identifying GRB10 rs933360.
Genome-wide quantile-quantile (Q-Q) (A) and Manhattan (B) plots for CIR in the meta-analysis. Regional plots of identified GRB10 genome-wide significant association for CIR in men (C) and women (D). Directly genotyped and/or imputed SNPs of GRB10 are plotted with their meta-analysis p-values (as −log10 values) as a function of genomic position (NCBI Build 36). In each panel, the strongest associated SNP is represented by a purple diamond (with meta-analysis p-value). Estimated recombination rates (taken from HapMap) are plotted to reflect the local linkage disequilibrium structure around the associated SNPs and their correlated proxies (according to a blue to red scale from r2 = 0 to 1, based on pairwise r2 values from HapMap CEU). Gene annotations were taken from the University of California, Santa Cruz, genome browser, as implemented in LocusZoom [41].