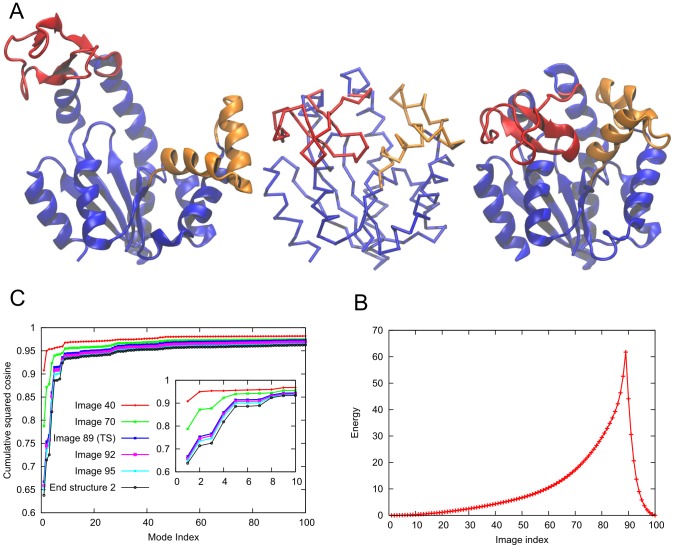
Figure 1. Conformational transition between the open and the closed states of adenylate kinase.
A. Structures of the open (left, PDB ID: 4AKE) and the closed (right, PDB ID: 1AKE) states. The LID and the NMP domains are shown in red and orange respectively. The CORE domain and the rest of the protein are shown in blue. The central structure is the 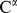 trace of the transition state produced by ANMPathway. B. The energy of the system along the transition. Total number of images in the pathway is 100, RMSD between two consecutive images is ∼0.1 Å. The transition state corresponds to image 89. C. Cumulative squared cosines between ANM modes and the change in structure between the initial state and a few selected conformers (images) along the transition pathway. The modes were calculated for the starting structure (open state). Here and in the counterparts generated for other test cases, the lowest frequency (slowest modes) end of the graph is enlarged in the inset. The force constants and cut-offs for both the end-states were set to 0.1 kcal/(mol Angstrom) and 15 Å in all applications in the present study, and no energy offsets were used for either of the end structures.
trace of the transition state produced by ANMPathway. B. The energy of the system along the transition. Total number of images in the pathway is 100, RMSD between two consecutive images is ∼0.1 Å. The transition state corresponds to image 89. C. Cumulative squared cosines between ANM modes and the change in structure between the initial state and a few selected conformers (images) along the transition pathway. The modes were calculated for the starting structure (open state). Here and in the counterparts generated for other test cases, the lowest frequency (slowest modes) end of the graph is enlarged in the inset. The force constants and cut-offs for both the end-states were set to 0.1 kcal/(mol Angstrom) and 15 Å in all applications in the present study, and no energy offsets were used for either of the end structures.