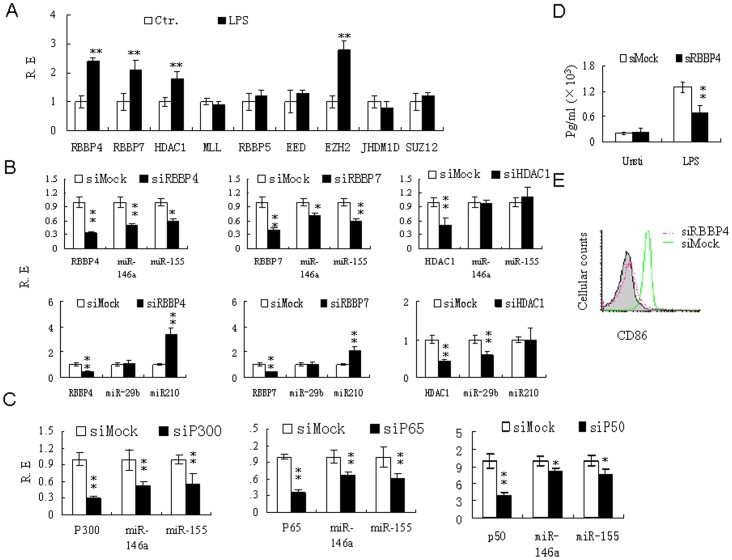
Figure 5. LPS associated factors affects the expression of miRNAs with H3K4me3 redistribution.
(A) qRT-PCR analyses of epigenetic factors RBBP4, RBBP7, HDAC1, MLL, RBBP5, EED, EZH2, JHM1D and SUZ12. Total RNA was extracted from moDCs following exposure to LPS (100 ng/ml) for 24 hrs and the expressions of RBBP4, RBBP7, HDAC1, MLL, RBBP5, EED, EZH2, JHM1D and SUZ12 were analyzed by qRT-PCR. (B) qRT-PCR analyses of miR-146a, miR-155, miR-29 and miR-210 in siRNA-transfected moDCs. MoDCs were transfected with siRNAs against RBBP4 (siRBBP4), RBBP7 (siRBBP7), HDAC1 (siHDAC1) or with control siRNA (siMock). After 48 hrs, total RNAs were extracted and analyzed by qRT-PCR. (C) qRT-PCR analyses of miR-146a and miR-155 in siRNA-transfected moDCs. moDCs were respectively transfected with siRNAs against p300 (siP300), p65 (sip65), p50 (siP50), or with control siRNA (siMock). After 48 hrs, total RNA was extracted and analyzed by qRT-PCR. (D) TNFα ELISA analyses of supernatants from siRBBP4 transfected moDCs. The supernatants were collected from siRBBP4 transfected moDCs and the TNFα concentrations were analyzed by ELISA (R&D, USA). (E) Flow cytometry analysis of CD86. siRBBP4 transfected moDCs were stained with anti-CD86 (IT2.2) and analyzed by flow cytometry. Grey, isotypic control. *, P<0.05; **, P<0.01.