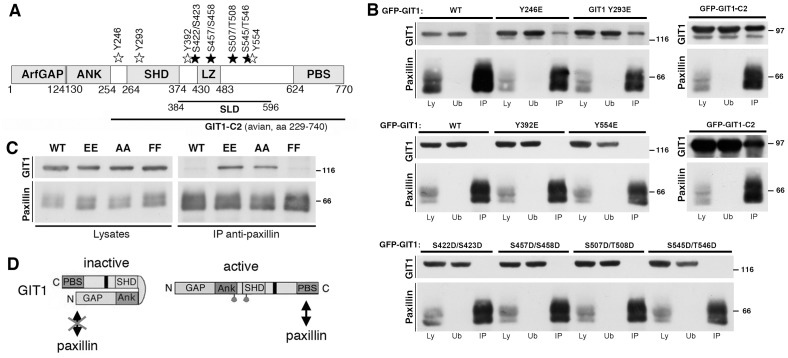
Figure 1. Mutations Y246E and Y293E of GIT1 enhance binding to paxillin.
(A) Schematic representation of human GIT1 (NP 001078923.1). GFP-tagged wild type GIT1 was used to introduce phosphomimetic mutations. Tyrosine (Y) or serine (S) and threonine (T) residues were mutated into glutamic acid or aspartic acid, respectively. White and black stars indicate the locations of the mutated tyrosine and serine/threonine residues, respectively. ArfGAP, Arf GTPase-activating protein; ANK, ankyrin repeats; SHD, Spa2-homology domain; LZ, leucine zipper; PBS, paxillin binding site; SLD, synaptic localization domain. (B) Aliquots of lysates (400 μg) from cells transfected with the indicated constructs were used for immunoprecipitation of endogenous paxillin. Filters with immunoprecipitates (IP), and equal amounts (80 μg) of the respective lysates (Ly) or unbound fractions after immunoprecipitation (Ub) were blotted with anti-GFP (for GIT1) or anti-paxillin antibodies. Molecular weight markers are indicated to the right of each blot. (C) The double substitution of residues Y246 and Y293 with either two glutamic acid (EE) or two alanine residues (AA) enhanced GIT1 binding to paxillin. Aliquots of lysates (200 μg protein) from cells transfected with the indicated constructs were immunoprecipitated with anti-paxillin antibody. Filters with immunoprecipitates (IP), and lysates (40 μg) were blotted as indicated, using anti-GFP (for GIT1) or anti-paxillin antibodies. (D) Model for GIT1 activation: see details in the text.