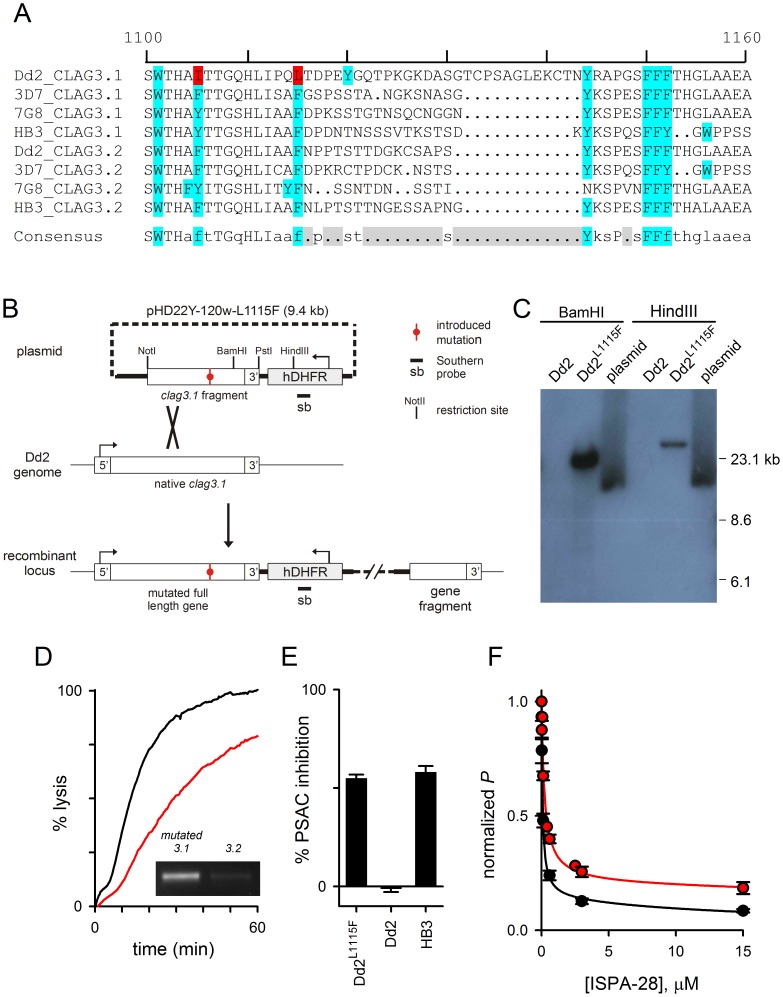
Figure 4. A variant extracellular residue accounts for PSAC resistance to chymotrypsin in Dd2 parasites.
(A) Multiple sequence alignment of an extracellular loop on indicated CLAG3.1 and CLAG3.2 sequences, representing geographically divergent parasites (Dd2, from Indochina; 3D7, probably Africa; 7G8 Brazil; HB3, Honduras). A variable segment is apparent in gray shading (Consensus); residues susceptible to chymotrypsin cleavage are shown in blue. Two sites refractory to cleavage in Dd2 CLAG3.1 are highlighted in red. (B) Schematic showing the allelic exchange strategy to introduce a single mutation in the Dd2 clag3.1 gene. Plasmid carrying the mutation is shown at the top; single homologous recombination into the Dd2 genome produces an intact full-length gene with a single site mutation and unchanged UTR sequences. (C) Southern blot showing integration of plasmid into the Dd2L1115F genome. Indicated DNA samples were digested and probed with an hDHFR-specific probe. Dd2 is not recognized by this probe, but Dd2L1115F yields a single band whose size differs from that of the plasmid. (D) Osmotic lysis kinetics for Dd2L1115F without and with 1h chymotrypsin treatment (black and red traces, respectively). Inset shows preferential expression of the mutated clag3.1 gene in this clone. (E) Mean ± S.E.M. chymotrypsin-induced inhibition for indicated parasites. (F) Dose responses for inhibition of sorbitol permeability (P) by ISPA-28. Black and red symbols represent mean ± S.E.M. inhibition for Dd2 and Dd2L1115F parasites, respectively. Solid lines represent best fits to the sum of two Langmuir isotherms.