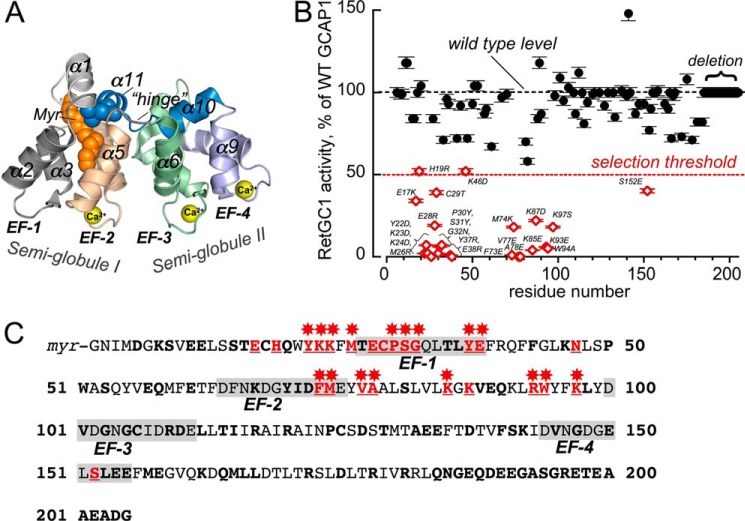
FIGURE 1.
Effect of amino acid substitutions in GCAP1 on RetGC1 activation. A, three-dimensional model of Ca2+-liganded GCAP1 (24) annotated as follows: Myr, N-myristoyl moiety; EF-1–EF-4, EF-hand domains; α1–α11, α-helices numbered beginning from the amino terminus; hinge, the loop connecting two semi-globules (I and II) between the α5 and α6 helices. Ca2+ ions bound in three metal-binding EF-hand loops (EF-2 through EF-4) are shown as yellow spheres. B, RetGC1 activation by 5 μm GCAP1 mutants normalized to the wild type activation in control samples (mean ± S.D., n = 3). The following mutations were tested: K8E, S9R; E11,12K; S15R,T16A; E17K,C18D; C18T,C106T,C125T; H19R; Q20R; Y22D; K23D; K24D; M26R; T27K; T27E; E28R; C29T; P30Y; S31Y; G32N; Q33R; T35R; L36E; Y37R; E38R; Q41R;; K46D; N47R; P50G; W51N,S53R; E57R; Q58R; E61R; F65N; K67D; Y70A; F73E; M74K; V77E; A78E; S81A; L82S; K85E; K87D; V88R; E89R; Q90R; R93E; W94A; K97S; V101Y; G103R; C106D; R109D; D110R; R117D; R120D; D127R; A132R; E133R; E134R; D137R; F140A; S141Y; K142D; V145R; G147R; E150Y; S152E; L153R; E154C; E155G; M157R; E158R; K162E; Q164R; L166R; L167R; R172E; D175K; R178D; R181E, Q184R; and deletion, ΔGln-184—Gly-205. Additional substitutions, I122E, N1123A, P124E, C125Q, S126Q, D127G,S128K, T129L, M130L, T138R, S141L, and V145E, and the Val-160–Gly-205 region replacement with the corresponding region from GCAP2 were tested as a single chimera construct (47). The assay contained 10 mm MgCl2 and 2 mm EGTA. The threshold level of 50% activation (dashed line) was selected for segregating the mutants for suspected damage of the RetGC1-binding interface. The mutations that caused this decrease are shown in open diamonds, and the substitutions are labeled next to the data points. C, positions of the mutations causing major decrease in RetGC1 activating capacity in the GCAP1 primary structure. All mutated side chains are marked in bold; those in which replacement rendered RetGC1 activation ≤50% of the wild type level are marked in red and underlined, and those in which mutations reduced activation below 80% are marked with red asterisks. The 12-residue loops of EF-1 through EF-4 are shaded.