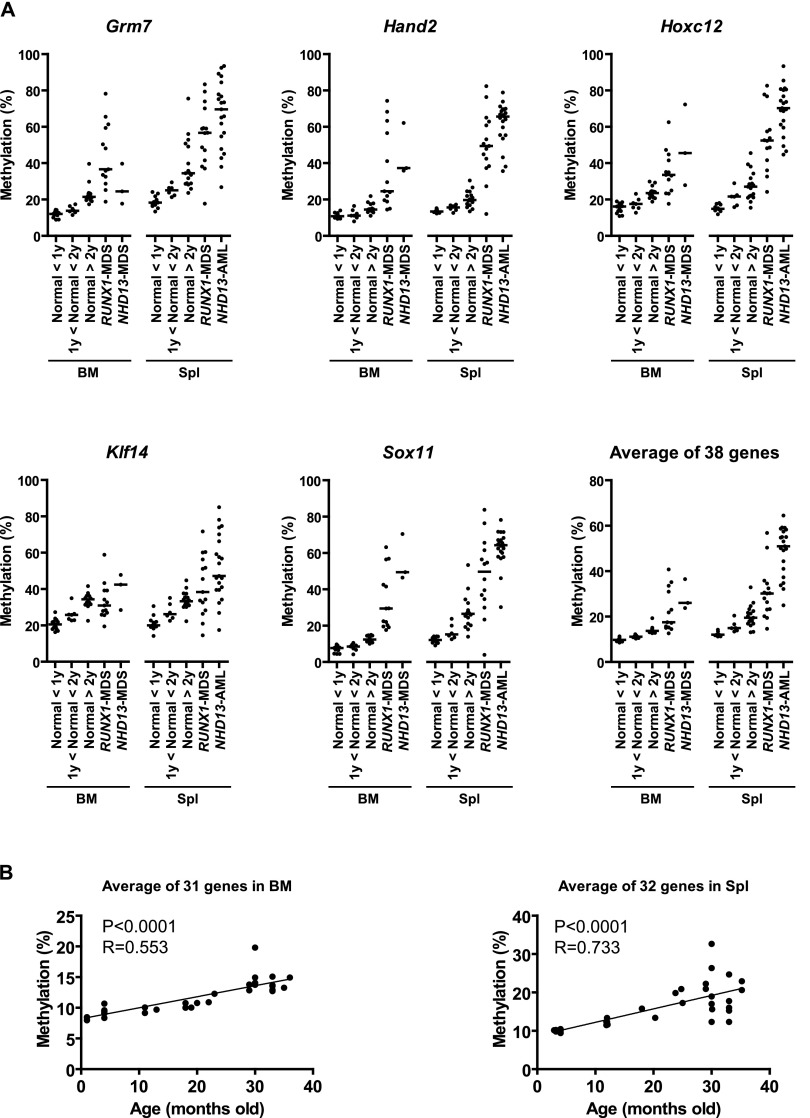
Figure 2.
DNA methylation profiles by bisulfite pyrosequencing analysis. (A) DNA methylation analysis in MDS mice. The percentages of methylated cytosines in the samples as obtained from pyrosequencing analysis. Each dot corresponds to one animal. We show here data on Grm7, Hand2, Hoxc12, Klf14, and Sox11. The other 33 hypermethylated genes are shown in Supplemental Figures S1 and S2. The averaged data were derived from the methylation data of 38 genes prone to hypermethylation. The bar in the graphs represents the median. (B) Methylation profiles of hypermethylated genes with age in mouse bone marrow and spleen. Association of the averaged percentages of methylated cytosines in the samples as obtained from pyrosequencing (y-axis) with age (x-axis) for significant genes.