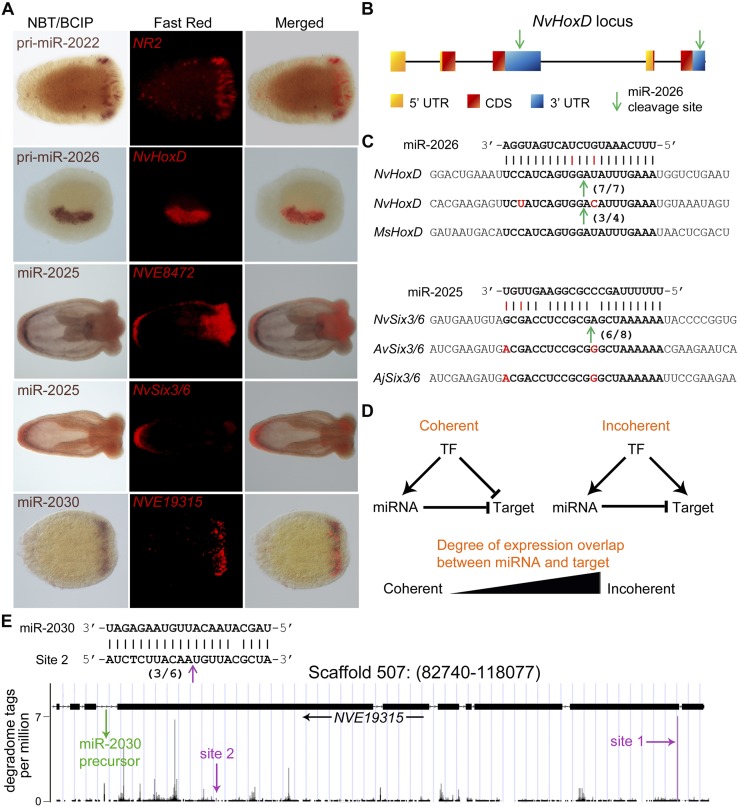
Figure 5.
Regulation of miRNA targets and target conservation. (A) Expression of microRNAs and their targets as determined by double in situ hybridization. (B) A scheme showing the structure of the partially duplicated HoxD gene and the position of the two binding sites of miR-2026. (C) The duplicated 3′ UTRs of HoxD in Nematostella (Nv) are very derived, yet the two binding sites of miR-2026 are highly conserved, and both transcript variants are cleaved. The site is also conserved in the sea anemone Metridium senile (Ms). Similarly, the miR-2025 binding site in the coding sequence of the Six3/6 transcript is conserved in Nematostella, Anemonia viridis (Av), and Anthopleura japonica (Aj). In panels B and C, cleavage positions are indicated by an arrow, and ratios of positive clones appear in brackets. (D) Schematic representation of coherent and incoherent circuits involving a transcription factor (TF) that regulates the expression of a miRNA and its targets and the degree of overlap between the expression domains of a miRNA and its targets that would indicate each of the two topologies. (E) miR-2030 and its host/target gene NVE19315 represent a clear case of incoherent regulatory circuit. The precursor of miR-2030 is located in an intron of NVE19315 as indicated by the green arrow. miR-2030 targets two sites in the NVE19315 transcript (indicated by magenta arrows) as supported by the degradome analysis (Site 1) and RLM-RACE (Site 2; cleavage position is indicated by an arrow, and ratios of positive clones appear in brackets).