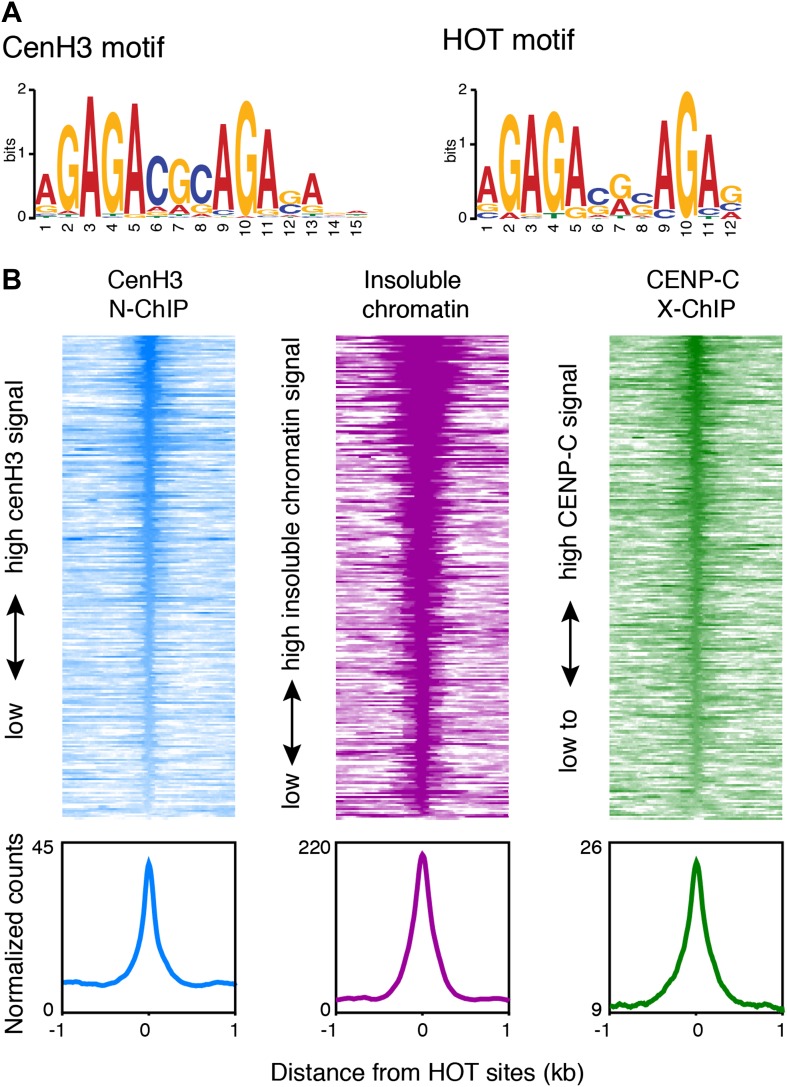
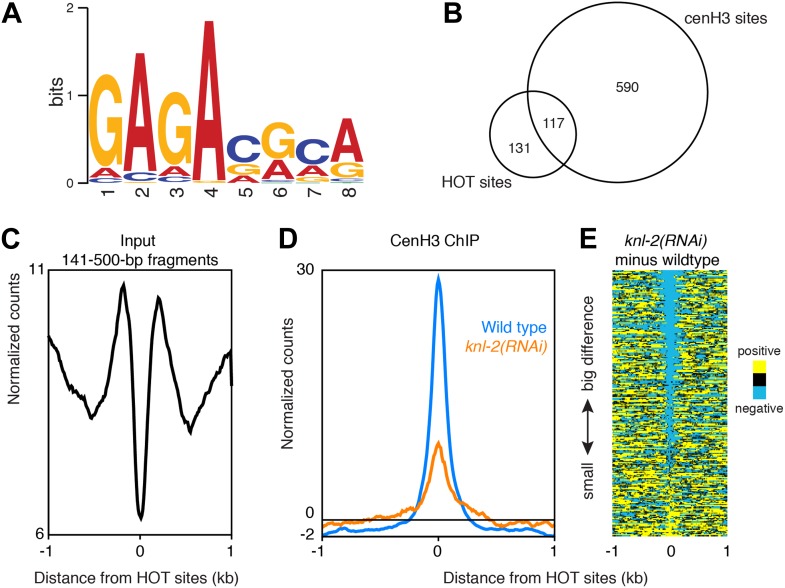
Figure 6. Centromeres coincide with transcription factor hotspots.
(A) MEME motif for the 80-bp cores of centromeric sites (left) and the 80-bp cores of high occupancy target (HOT) sites (right). (B) Heatmaps and average plots of cenH3 ChIP, insoluble chromatin and CENP-C ChIP signal within a 2-kb window around HOT sites, illustrating that HOT sites are highly occupied by cenH3, insoluble chromatin and CENP-C. Each line of the heatmaps represents an individual HOT site. Heatmaps are sorted by decreasing signal.