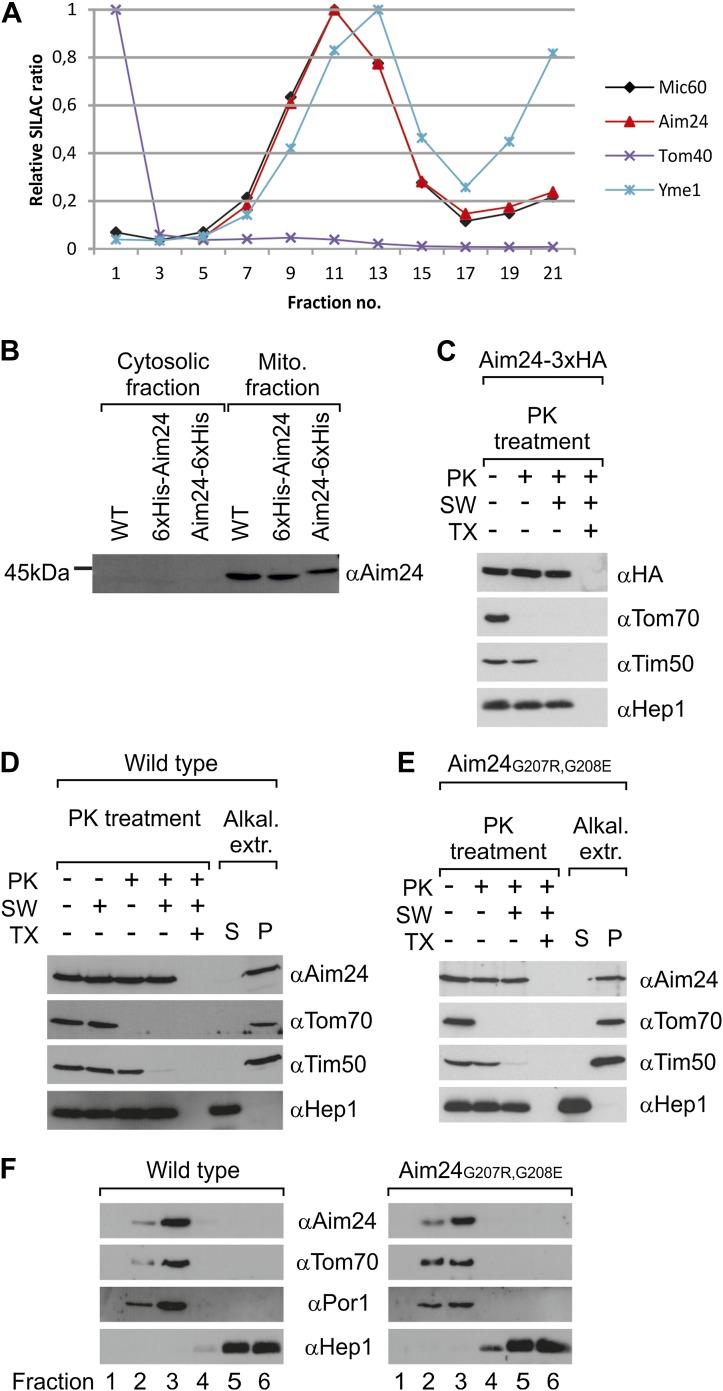
Figure 1. Topology of Aim24.
(A) Aim24 is a contact site protein. Isolated mitochondria from wild type (YPH499) cells were subjected to osmotic shrinking, sonication and buoyant density fractionation. Gradient fractions were analyzed by SILAC supported quantitative mass spectrometry (Harner et al., 2011). The distribution of Aim24 in the gradient is shown in red. Distribution of marker proteins of the OM (Tom40), the IM (Yme1) and of the contact site protein Mic60 are included. (B) Aim24 is synthesized as a precursor with an N-terminal targeting signal which is cleaved upon import. Wild type cells harboring pYES2 empty vector, Δaim24harboring pYES2 6xHis-Aim24 or pYES2 Aim24-6xHis were grown on SLac medium containing 0.1% glucose. Protein expression was induced by incubation for 30 min in SLac medium containing 0.1% galactose. Mitochondrial and supernatant fraction were analyzed by SDS-PAGE and immunoblotting with antibodies against Aim24.(C) The C-terminal HA-tag of Aim24-3xHA is not accessible to proteinase K (PK) added to intact mitochondria and mitoplasts. Isolated mitochondria were subjected to PK treatment, osmotic swelling (SW) and Triton X-100 (TX) as indicated and analyzed by immunoblotting. (D) Aim24 behaves like an integral membrane protein. Left: mitochondria isolated from wild type cells were treated as described in (C). Right: mitochondria were exposed to alkaline treatment at pH 12. Soluble (S) and membrane integrated (P, pellet) material was separated by centrifugation and analyzed by immunoblotting. (E) Exchange of two glycine residues in a hydrophobic stretch of Aim24 by charged residues does not alter its firm association with the membrane. Mitochondria were isolated from a Δaim24 strain harboring the pYES2 plasmid encoding Aim24 G207R, G208E and treated as described in (C). (F) Neither Aim24 wild type nor Aim24 G207R, G208E protein aggregate upon alkaline treatment of mitochondria. Mitochondria were subjected to alkaline treatment and proteins were separated by flotation gradient centrifugation. The gradients were fractionated (fractions 1–6), proteins were TCA precipitated and analyzed by immunoblotting. Fraction 1 is top and fraction 6 is bottom.