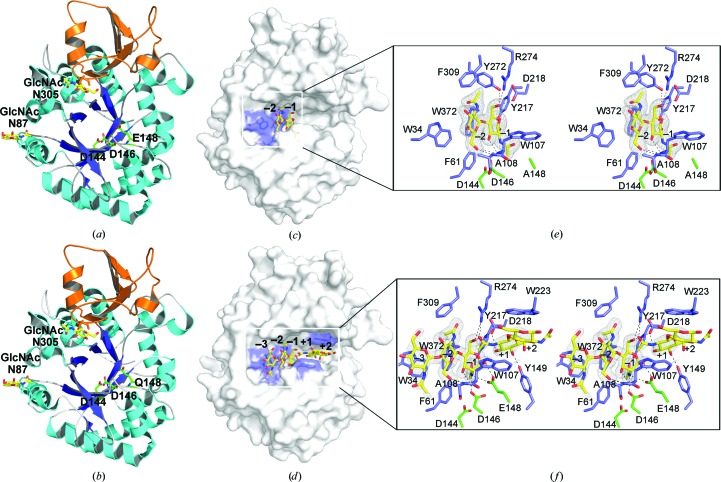
Figure 1.
Overall structures of unliganded and oligosaccharide-complexed OfChtI-CADs. (a, b) Cartoon representation of OfChtI-CAD (a) and the E148Q mutant (b). The structure consists of two domains: a core domain with an (α/β)8 TIM-barrel fold (cyan, α-helices; blue, β-strands) and an insertion domain (orange). The catalytic residues and the N-GlcNAc residues at the N-glycosylation sites are shown as sticks with green and yellow C atoms, respectively. (c, d) Surface representations of E148A complexed with (GlcNAc)2 (c) and OfChtI-CAD complexed with (GlcNAc)2/3 (d). The ligand is shown as a stick with yellow C atoms. The aromatic residues that stack with the sugar rings are shown in blue. The numbers indicate the subsite to which the sugar is bound. (e, f) Stereoview of the substrate-binding cleft with details of the interactions between (GlcNAc)2 and E148A (e) and between (GlcNAc)2/3 and OfChtI-CAD (f). The ligand is represented as a stick with yellow C atoms and the 2F o − F c electron-density map around the ligand is contoured at the 1.0σ level. The catalytic residues and the amino acids that interact with the ligand are labelled and are shown as sticks with green and blue C atoms, respectively. The numbers indicate the subsite to which the sugar is bound. Hydrogen bonds are drawn as dashed lines.