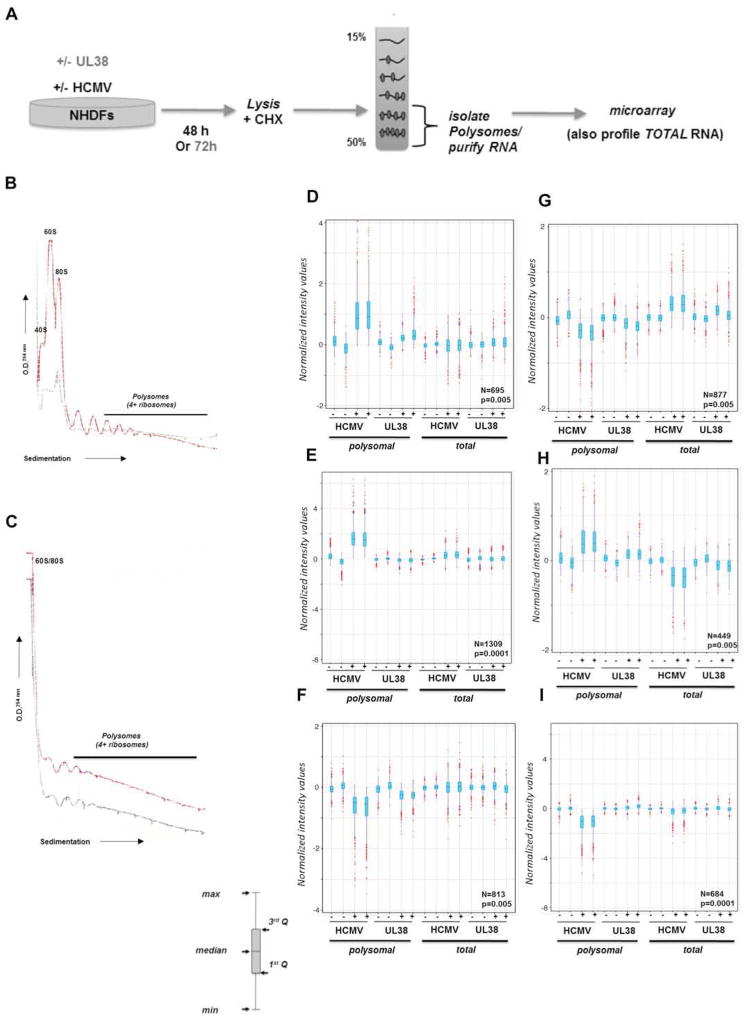
Figure 1. Polysome profiling reveals extensive changes to ongoing host mRNA translation in response to HCMV infection.
A) Illustration depicting experimental procedure. NHDFs (+/− HCMV or transduced with a lentivirus that expresses the HCMV UL38 ORF from a dox-inducible promoter) were lysed in the presence of cycloheximide (48 hpi or 72 h post-induction of UL38). Extracts were sedimented through a 15–50% linear sucrose gradient that was subsequently fractionated and absorbance at 254 nm monitored. RNA isolated from fractions containing 4 or more ribosomes was used to prepare cRNA probes and hybridized to a DNA microarray. Total RNA isolated from parallel cultures was used to prepare cRNA probes and hybridized to a microarray to identify overall mRNA abundance changes in response to HCMV infection or UL38 induction. B) Absorbance tracing (254 nm) comparing mock-infected (gray line) vs HCMV-infected (red line) cultures. Migration of ribosome subunits (40S, 60S), monosomes (80S), and polyribosomes (4 or more ribosomes) are indicated. The top of the gradient is on the left. C) As in B but using uninfected cultures stably transduced with a doxycycline (dox)-inducible UL38-expression vector in the absence (gray line) or presence (red line) of dox. While polyribosome peaks were detected in uninfected cells, signals from 40S / 60S subunits and 80S monoribosomes were not resolved in the linear range of the tracing. D–I) Boxplot graphs based on normalized intensity values are shown with corresponding probe number (N) and Pavlidis Template Matching (PTM; Saeed et al., 2003) p-value indicated in the lower right corner. Outliers (in red), whiskers (indicating maximum and minimum), interquartile range (light blue shading) and median are shown. PTM algorithm (see methods) performed at p-values ranging from 5 × 10−3 to 10−5 identified specific profile patterns for groups of mRNAs in duplicate samples of polysomal and total RNA (−/+ HCMV; −/+ UL38 induction) that were elevated both in the polysomal fraction of HCMV-infected and dox+ (UL38-induced) cells (D); mRNAs that were elevated only in the polysomal RNA fraction from HCMV-infected cells (E); mRNAs reduced only in polysomal fraction from HCMV-infected and dox+ (UL38-induced) cells (F); mRNAs reduced in both polysomal RNA fraction from HCMV-infected and dox+ (UL38-induced) cells but elevated in the total RNA from the same conditions (G); elevated in both polysomal RNA fractions from HCMV-infected and dox+ cells but reduced in the total RNA from the same conditions (H); reduced only in polysomal RNA from HCMV-infected cells (I).