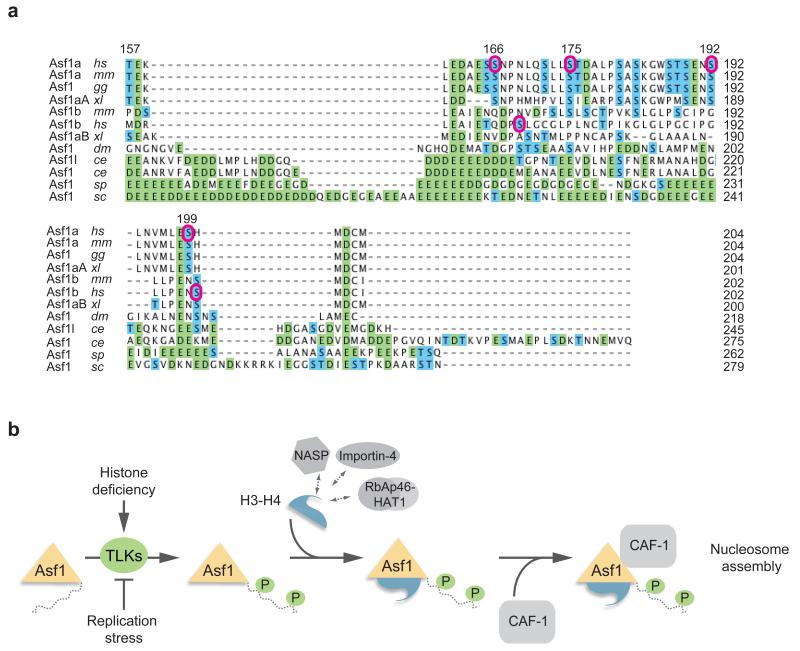
Figure 7. Phosphorylation site conservation and model.
(a) Sequence alignment of the Asf1 C-terminal tails performed by the ClustalW2 algorithm. Homo sapiens, hs; Mus musculus, mm; Gallus gallus, gg; Xenopus laevis, xl; Drosophila melanogaster, dm; Caenorhabditis elegans, ce; Saccharomyces cerevisiae, sc; Schizosaccharomyces pombe, sp. Acidic residues are indicated in green and Ser/Thr residues are in blue. Purple circles denote TLK1 phosphorylation sites in human Asf1a and Asf1b identified in this study. (b) Hypothetical model for how TLK phosphorylation fine-tunes Asf1-dependent histone provision during genome duplication. TLKs bind histone-free Asf1 and phosphorylate the tail at multiple sites. This in turn facilitates complex formation with histones and the downstream chaperones HIRA and CAF-1. TLK signaling responds to histone demand; Asf1a phosphorylation increases when cells lack new soluble histones and decreases if DNA synthesis is blocked and histones demand drops.