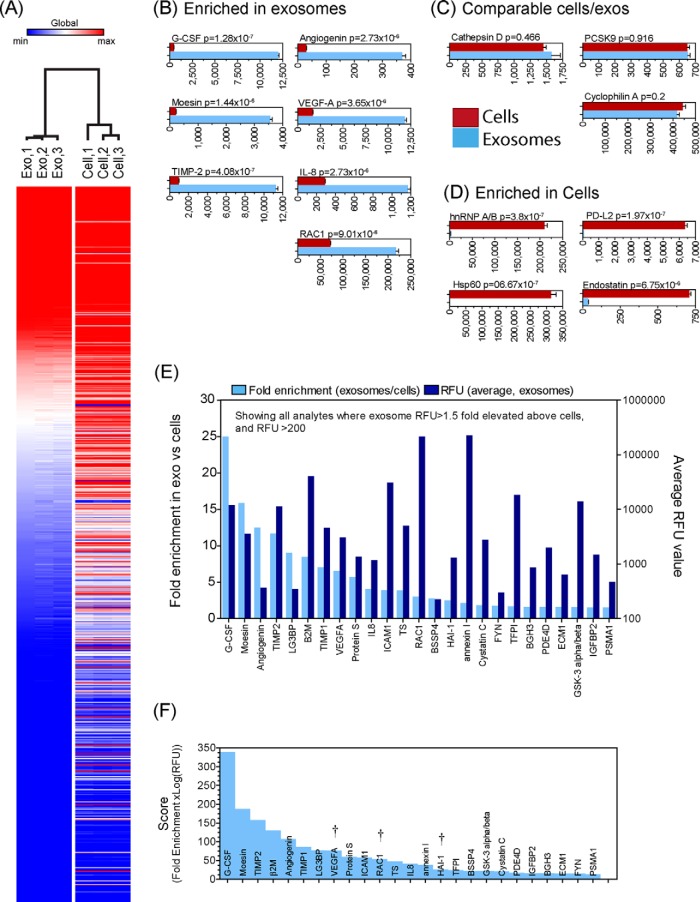
Fig. 3.
Comparison of exosomes and cells under SB17 conditions. Exosomes and cell preparations under SB17 conditions were compared, and the data, filtered to eliminate poor replicates, are represented as a heat map highlighting the dissimilarity between exosomes and their parent cells (A). A total of 33 proteins demonstrated 1.5-fold or greater increased expression in exosomes relative to cells, and some examples of these are shown (bars show mean RFU ± S.E. of triplicates, with specified p values) (B). 24 proteins exhibited comparable expression levels (of less than 0.1-fold difference) (C), whereas 574 proteins were >1.5-fold more abundant in cells (D). The data were filtered to include only analytes reporting >1.5-fold elevation in exosomes and those with an RFU signal of >200 units, and these are shown (E), plotted according to fold enrichment (left axis) and RFU values (right axis). A simple multiplication of fold-increase × log2(RFU), used as a means of identifying proteins that may be both enriched and highly abundant in exosomes, is shown (F). Some candidate proteins were selected from this plot for subsequent validation analyses, and these are indicated with †.