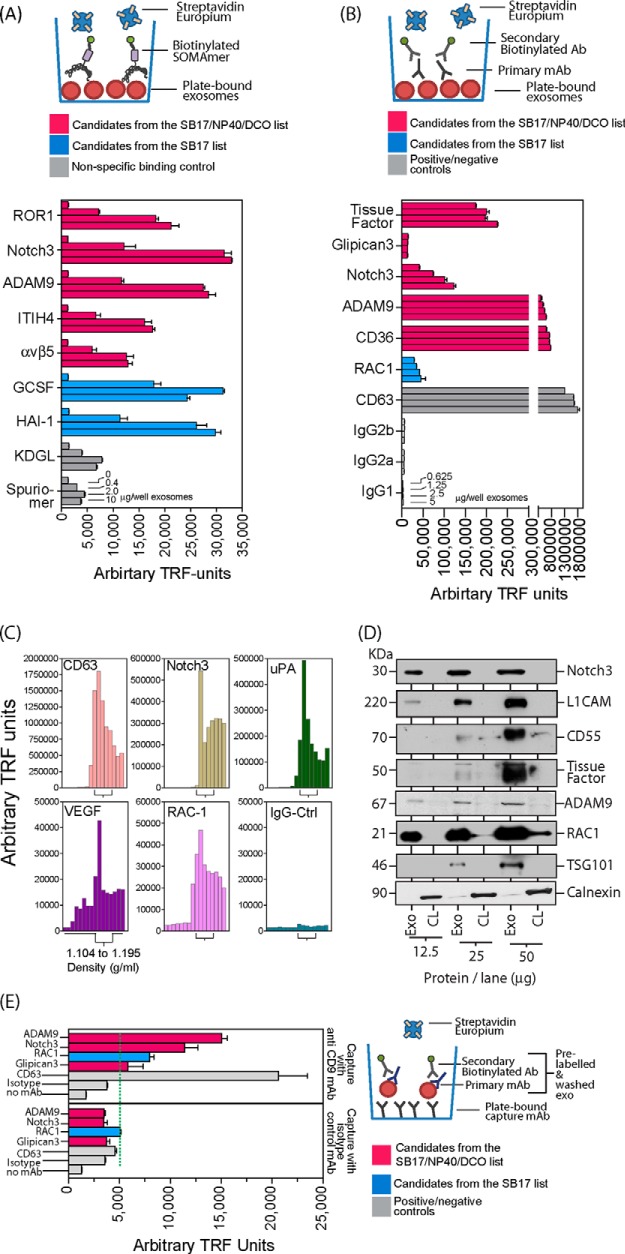
Fig. 4.
Confirmation of the expression of selected SOMAscanTM-identified proteins. Du145 exosomes purified via the sucrose cushion method were immobilized at specified doses on ELISA microplates and probed using individual SOMAmers® (A) or an indirect staining method with antibodies (B). Signals were detected using Europium-streptavidin and time-resolved fluorimetry (TRF) in each case (bars represent mean ± S.E. of duplicate measurements). SOMAmers® (KDGL or Spuriomer) act as irrelevant controls for nonspecific binding (A). To confirm that the identified proteins float at a classical exosomal density, a continuous sucrose gradient fractionation was performed, and the density of collected fractions was determined prior to immobilization on microplates as described above. Proteins were detected with antibodies using the same indirect staining method as described above. The fractions with densities between 1.1 and 1.2 g/ml are annotated (C). Whole cell lysates and exosomes normalized for protein were subjected to SDS-PAGE and Western blotting and probed with antibodies as indicated. This revealed relative exosomal enrichment for all candidates, whereas calnexin exhibited the reverse pattern (D). Sucrose cushion-purified exosomes were labeled in solution with primary antibody (as specified) and secondary biotinylated antibody before being immobilized on microplates pre-coated with anti-CD9 or isotype control antibody. After washing, signals were detected using Europium-streptavidin and TRF (bars represent mean ± S.E. of quadruplicate measurements) (E).