Fig. 1.
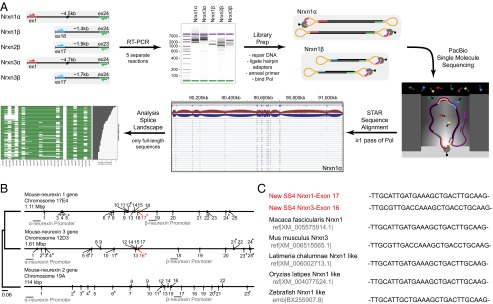
Single-molecule long-read sequencing of full-length neurexin mRNA transcripts. (A) Schema of the workflow of Pacific Biosciences sequencing of full-length neurexin transcripts for mapping neurexin diversity. Full-length transcripts of Nrxn1α, 1β, 2β, 3α, and 3β were separately reverse-transcribed and PCR-amplified (RT-PCR) from total RNA isolated from adult murine prefrontal cortex. PCR was performed with primer pairs specific to the first and last coding exons of each gene, with α- and β-specific the forward primers and identical reverse primers for the α- and β-versions of a given neurexin. The cDNA size distribution for each neurexin was examined by gel electrophoresis before preparation of Pacific Biosciences sequencing libraries by blunt-end ligation of hairpin adapters, annealing of the sequencing primer, and binding of biotinylated DNA polymerase. Subsequently, full-length transcripts were sequenced by single-molecule long-read sequencing (36), yielding an average read length of 3.6–5.8 kb. Reads containing at least two adapters (≥1 passes through circular DNA by the polymerase) were processed (quality filter, adapter removal) and then aligned to the mouse genome (mm10) using the STAR aligner (38) and used for further analysis. (B) Phylogenetic tree and structures of mouse neurexin genes. The diagrams depict the positions of exons and introns. Exons are numbered; asterisks mark exons subject to canonical alternative splicing. The positions of α- and β-neurexin promoters are indicated, and the position and size of the genes are shown above each gene diagram (see also Fig. S1). (C) Neurexin mRNAs from diverse species contain the newly identified SS#6. Sequences of murine Nrxn1α and Nrxn3α SS#6 that were identified in this study are colored in red.
