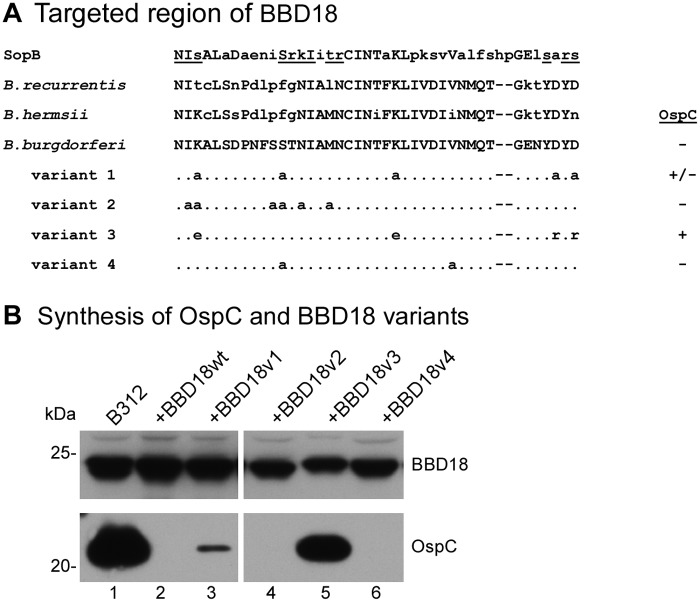
FIG 3 .
OspC regulatory activity of BBD18 site-directed mutants. (A) Alignment of amino acid residues 178 to 220 of the SopB helix-turn-helix motif, residues 52 to 92 of the BBD18 homolog in the relapsing fever spirochetes B. recurrentis and B. hermsii, and residues 56 to 96 of WT B. burgdorferi BBD18, with the same region of the four BBD18 variants (v1 to v4) with site-directed mutations. The underlined residues of the SopB sequence are those that have been shown to contact DNA (35). Amino acid residues that differ from the WT B. burgdorferi BBD18 sequence are shown in lowercase letters. Residues that have been changed in BBD18 variants are specified, whereas unchanged residues in this region are represented with dots; the remainder of the BBD18 sequence not depicted in this alignment is identical between the WT and variant BBD18 proteins. Synthesis of OspC by spirochetes coexpressing WT or variant bbd18 alleles (as shown in panel B) is indicated to the right of the alignment. (B) BBD18 (top) and OspC (bottom) immunoblots of B312 harboring WT or variant bbd18 alleles (variant 1 to variant 4 in panel A) expressed from the flaB promoter on a shuttle vector. Whole-cell lysates of B312 (lane 1), B312/pBSV2*flaBp-bbd18 (+BBD18wt, lane 2), and B312 harboring the bbd18 variants (+BBD18v1 to -v4, lanes 3 to 6) were analyzed by immunoblotting with antiserum recognizing BBD18 or OspC, as indicated. Lanes 1 to 3 and 4 to 6 were from nonadjacent regions of the same immunoblots. The molecular mass (kDa), based on the mobility of protein standards, is denoted on the left, and lane numbers are indicated beneath the panel.