Figure 1. Properties of VEGF isoforms and proteolytic cleavage sites.
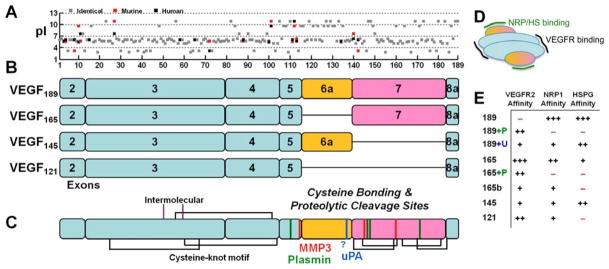
A, The acidity of the individual amino acids (pI) for human (black) and murine (red) VEGF shows the basic residues responsible for the heparin-binding domains of exons 6 and 7. Murine VEGF contains a deletion of Gly-8 found in human VEGF and is frame-shifted for comparison. The overall sequence identity between murine and human VEGF189 orthologs is 89%. B, Exon structure of the predominant VEGF isoforms in humans, scaled to the 189 amino acids shown in panel A. Note that VEGF165/164 replace the last residue in exon 5, Lys, with Asp. Exon 1 is not present in processed VEGF, it is removed by signal peptidase. Of the anti-angiogenic VEGFxxxb isoforms, which use exon 8b instead of 8a, VEGF165b is most common. C, Disulfide bonding structure (black and purple lines) (223) and known proteolytic cleavage sites for the serine proteases (plasmin, green; uPA, blue) and MMPs (red). The uPA cleavage site has not been specifically mapped but is thought to reside in the C-terminal portion of exon 6a. Exon 7 is linked to the first amino acid in exon 8 in all isoforms except VEGF121. Processing of exon 7 by the MMPs results in a single fragment due to linkage by disulfide bonds, whereas complete processing by plasmin may yield a second two-amino-acid Arg147-Lys148 (in VEGF189) fragment. D, VEGF receptors (VEGFR1, R2, R3) bind to the bivalent dimeric ligand in the region encoded by exons 3 and 4 (blue). Neuropilins (NRP) and heparan sulfates (HS) bind VEGF in the region encoded by exons 6 and 7 (yellow/purple). E, VEGF isoforms differentially bind to Neuropilin-1, VEGFR2, and heparin/heparan sulfate. Cleavage by plasmin (P) and uPA (U) can activate VEGFR2 binding in VEGF189 and decreases NRP1 and HSPG binding.
