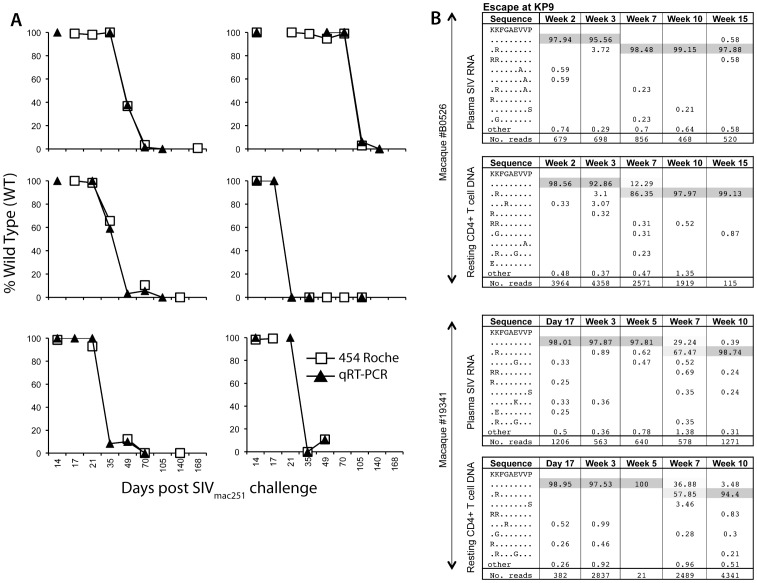
Figure 1. Analysis of escape at the KP9 Gag CTL epitope by pyrosequencing.
A. Estimation of K165R KP9 escape in serial resting CD4+ T cell SIV DNA samples using pyrosequencing compared to KP9-specific qRT-PCR. Six representative macaque examples comparing CTL escape at KP9 from serial resting CD4+ T cell SIV DNA samples after infection with SIVmac251 determined using the KP9-specific qRT-PCR compared to pyrosequencing. B. KP9 escape in plasma SIV RNA and resting CD4+ T cell SIV DNA for 2 representative macaques using Roche 454 sequencing. Examples of KP9 CTL escape in plasma SIV RNA and resting CD4+ T cell SIV DNA 2 animals using pyrosequencing. The CTL amino acid sequence is shown in the first column, with the % of sequence in the subsequent columns and the time point post SIV challenge at the top of the column. The mutation identified is shown at each time point with the total reads shown in the bottom row. Common variants at each time point are shaded with rarer variants accounting for the remaining sequences.