Abstract
Members of the Diaphanous (Dia) protein family are key regulators of fundamental actin driven cellular processes, which are conserved from yeast to humans. Researchers have uncovered diverse physiological roles in cell morphology, cell motility, cell polarity, and cell division, which are involved in shaping cells into tissues and organs. The identification of numerous binding partners led to substantial progress in our understanding of the differential functions of Dia proteins. Genetic approaches and new microscopy techniques allow important new insights into their localization, activity, and molecular principles of regulation.
Keywords: Drosophila, cytoskeleton, actin, nucleator, development, formin
Introduction
Cells evolved different actin nucleators that catalyze the nucleation reaction, the rate-limiting step in actin polymerization. Diaphanous (Dia) was originally identified by its essential function in cytokinesis1 and constitutes a major branch of the formin family, which together with Arp2/3 and WH2 proteins represents the majority of actin nucleators in cells.2 The formin protein family is defined by the presence of the formin homology domain 2 (FH2)3 and is further classified according to the presence and arrangement of additional domains.4 Dia proteins contain a RBD/FH3 region in their N-terminal part, which consists of a Rho-binding domain (RBD), 4 Arm repeats (also called Dia-inhibitory domain, DID), a dimerization domain (DD), and a putative coil-coiled region (CC, Figure 1). Phylogenetic sequence analysis suggests that the emergence of Dia predates the evolution of animals and thereby multicellularity.
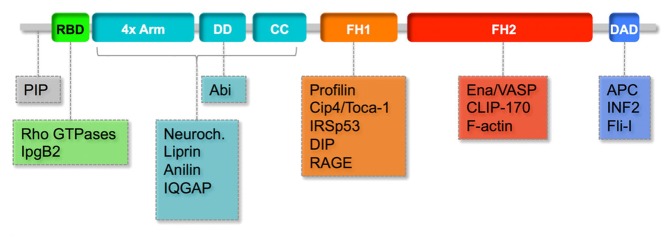
Figure 1. Domain structure and interactors of Dia. Dia consists of the formin homology domains 1 and 2 (FH1, FH2), Rho-binding domain (RBD), 4 Armadillo repeats (4x Arm, also called Dia inhibitory domain, DID), dimerization domain (DD), a predicted coiled-coil region (CC) and a dia autoregulatory domain (DAD). 4x Arm, DD, and CC are also referred to as FH3. Structure is shown for mDia1 (uniprot O08808,41). Dia interacting proteins are associated with the domains and regions they bind to.
Revisiting the evolutionary history of Dia
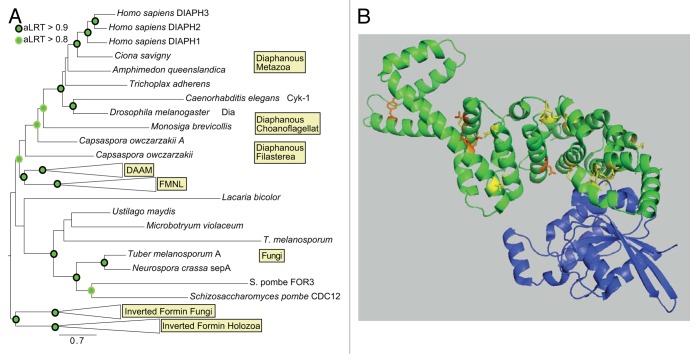
Analyses of the evolution of Dia have mostly been embedded in larger studies considering the complete formin family and thus focused on their common denominator, the FH2 domain. The position of the Dia subfamily varies in previous publications. In the initial phylogenetic classification by Higgs and Peterson, 7 metazoan groups were defined, which were separate from the formins in fungi, plants, and protists. Based on available non-FH2 sequences, Dia, DAAM, and FMNL subfamilies were proposed to be grouped together.3,5 The position of non-yeast fungi was not resolved, and the yeast formins were the outgroup of all taxonomic groups including plants. Rivero et al. resolved the formin subfamilies but did not find sufficient bootstrap support for defining the relationship among them.6 Out of the fungi genomes, only the yeast formins were included, which built a clade on their own. As a first evidence for a relationship among the subfamilies, Chalkia et al. grouped the FMNL and the DAAM subfamily together, when considering animals and choanoflagelates.7 By focusing only on metazoan species, the clade consisting of FMNL and DAAM was further corroborated, with Dia and the inverted formins as sister clade. A first hint of a possible older evolutionary origin of Dia was shown by Grunt et al., who identified a protein from Monosiga brevicollis, which grouped together with metazoan Dia.8 As in other studies, non-yeast and yeast fungal sequences formed their own clade. Interestingly, a phylogenetic tree based on the domains beside the FH2 supported a clade with DAAM and FMNL proteins.6
To revisit the relationship of the formin subfamilies and to define the position of the Dia subfamily (Fig. 2A), we combined the sequences of the N-terminal RBD/FH3 region common to the Dia subfamily (RBD, 4xArm/DID, DD, CC, Figure 1) and the FH2 domain to increase the phylogenetic resolution. Our analysis was therefore restricted to the Dia, DAAM, FMNL, and inverted formin subfamilies, as they are the only members containing a RBD/FH3 region. We extracted genes with this domain architecture from selected fungal and holozoan genomes. In the following, we merged the domains as identified by SMART (the DID/DD region is covered by the Hidden Markov Models (HMMs) for Drf_GBD and Drf_FH3; a single HMM exists for the FH2 domain).9 Then, the sequences were aligned using MUSCLE10 and the phylogenetic tree was reconstructed using PHYML after identification of the best model with PROTTEST.11 In contrast to previous studies, we found a well_Supported clade containing the metazoan inverted formins and fungal sequences. Thus, we identified possible fungal orthologs of the inverted formins. With a supported origin predating the divergence of fungi and Holozoa, we used this clade as outgroup. As in previous studies, Dia, DAAM, and FMNL were reconstructed as well_Supported, monophyletic groups. Additionally, we were able to obtain support for the relationship between these subfamilies. Here, Dia is the oldest of the 3 subfamilies, and DAAM and FMNL evolved by a subsequent gene duplication. Within the Dia clade, the incorrect but well_Supported placement of the arthropod and nematode sequences might be attributed to long-branch attraction. Unexpectedly, also sequences from the chonaoflagellat Monosiga brevicollis and the filasterean Capsaspora owczarzakii were placed with high support within the Dia clade. Thus, the emergence of Dia predates the evolution of animals and thereby multicellularity. Contrasting previous analyses, we retrieved a single fungal clade, covering yeast and non-yeast proteins including Schizosaccaromyces pombe FOR3 and CDC12 with sufficient support. Although the position of this clade as outgroup to the holozoan Dia, DAAM, and FMNL is not reliably supported, it might indicate that these fungal genes comprise the orthologs of the holozoan families. In this scenario, the last common ancestor of ophistikonts contained a single gene, which evolved by holozoan specific duplications to Dia, FMNL, and DAAM.
Figure 2. The evolutionary origin of diaphanous. (A) Phylogenetic tree based on the RBD/FH3 region and the FH2 domain. Green circles denote an approximate Likelihood Ratio (aLRT) support of > 0.8, a black circle indicates aLRT > 0.9. For accession numbers of Dia sequences see Supplemental Materials. (B) Differentially conserved sites between vertebrate Dia paralogs. Positions with a zscore > 5 were mapped onto the structure of mDia1 (pdb: 3EG5; mDia1 in green, interacting Rho GTPase in blue). Sites conserved between Dia2 and Dia3 in yellow, differing sites in orange.
At the base of the vertebrates, further duplications gave rise to Dia1, 2, and 3. Usually, a gene retained after a gene duplication undergoes either neofunctionalization (i.e., one of the genes evolves a new function) or subfunctionalization (i.e., each gene retains a subset of the original function).12 In the case of Dia, one of the described functional differences is their affinity to small GTPases of the Rho family. mDia1 has a small number of interactors, whereas mDia2 and 3 are more promiscuous. Structural analyses revealed that this functional difference can be attributed to a small motif consisting of ‘NNN’ in mDia1, which is substituted to ‘TSH’ in mDia2 and 3.13 These positions are conserved throughout the vertebrate Dia1 and Dia2/3, respectively. This pattern is a showcase of class II functional divergence, where a position is conserved in both subfamilies, but harbors subfamily specific amino acids.14 As the existence of such sites might enable to home in on specificity determining sites, we aligned the mDia paralogues of different vertebrates und performed an SDPfox analysis15 (Table S1). Indeed, the already known motif was significantly identified (i.e., z-score < 5). Additionally, further positions, mostly in the N-terminus of Dia, were predicted. To estimate the relevance of these sites, we mapped them onto the structure of mDia1 (pdb:3EG5) (Fig. 2B).13 Indeed, further sites in addition to the NNN–TSH motif were predicted in proximity to the bound GTPase. This includes positions 210 and 262, which belong to the DAD interaction interface.16 Further sites were identified when comparing mDia2 and mDia3. This includes position 422, which harbors a tyrosine in mDia1 and 2 and a phenylalanine in mDia3. This position is part of the dimer forming area16 and might therefore be involved in the homodimerization of Dia proteins. The additional sites might be a good starting point for further analyses of functional differences between the Dia subfamilies. Interestingly, when comparing Dia2 and Dia3, 1 of the 2 kept the amino acid present in mDia1, whereas a substitution happened in the other (Table S1). This might indicate that both have retained parts of the functionality of Dia1.
Dia a multimodular actin regulator—a matter of the binding partner
Members of the Dia protein family are multimodular proteins that interact with numerous actin regulators, adapters, and signaling components (Table 1, Figure 1). Beside the formin homology domains FH1 and FH2, the presence of additional domains, i.e., RBD/FH3 and the C-terminal Dia-autoregulatory domain (DAD), is the characterizing feature of the Dia subfamily. The FH1 domain is proline rich and binds to a range of proteins. Most importantly, the FH1 domain binds Profilin,17-19 which is required for the elongation activity of Dia, albeit not for nucleation.20,21 Among the interactors of the FH1 domain are proteins with SH3 domains, such as the F-BAR protein Cip4/Toca-1, the I-BAR protein IRSp53, and the Dia interacting protein DIP/Wish.22-24 The F-BAR protein Cip4 contains an SH3 domain at its C-terminal end, which is necessary for colocalization of Cip4 and Dia in Drosophila S2 cells.22 This interaction is also involved in controlling Dia-catalyzed actin polymerization. Cip4/Toca-1 protein inhibits the Profilin-independent nucleation as well as the Profilin-dependent elongation. Single molecule analysis by TIRF microscopy revealed that about 90% of elongating filaments were suppressed by Cip4. However, about one-tenth of the filaments showed increased elongation speed, suggesting that these filaments escaped inhibition.22 The inhibition of nucleation and elongation by Cip4 seems to be mediated by the interaction of SH3 and FH1 domains, since the isolated SH3 domain was also inhibitory.22 The SH3 domain containing DIP inhibits actin polymerization in vitro,25 and interestingly also mDia2 dependent actin bundling.25-27 The inhibitory mechanism and the role of the SH3-FH2 interaction has remained unclear. The function of a third SH3 containing protein, IRSp53, on actin polymerization in vitro has not been reported yet. Given that the SH3 domain of Cip4 on its own showed inhibitory activity,22 it is likely that IRSp53 inhibits filament nucleation and elongation in a similar manner.
Table 1. Dia binding partners and their proposed functions.
| binding partner | binding domain | physiological function | biochemical function | references |
|---|---|---|---|---|
| Rho1/RhoA | RBD | activation of nucleation and elongation | release of autoinhibition | 13 , 16 , 19 , 41 , 63 , 110 |
| IpgB2 | RBD | 158 | ||
| PIP2, phospholipids | N-basic | apical/membrane localizaton | 47 - 49 , 158 , 161 | |
| Profilin | FH1 | required for elongation activity | 17 - 19 | |
| Cip4/Toca/FBP17 | FH1 | antagonises Dia mediated membrane stabilization and cytokinesis | inhibition of nucleation and elongation | 22 |
| IRSp53 | FH1 | filapodia induction | 24 , 81 , 162 | |
| DIP/Wish | FH1 | blebbing in amoeboid migratory cells | mDia2 dependent filament assembly and bundling | 23 , 25 , 163 - 165 |
| RAGE | FH1 | cell migration | 28 , 166 | |
| Ena/Vasp | FH2 | lamellipodia, SRF activity | 34 , 36 , 37 | |
| Clip-170 | FH2 | phagocytosis | 35 | |
| Neurochondrin | FH3 | unknown | 45 | |
| Abi | DD | junctions in MDCK cells | 44 | |
| Liprin | DID-DD | antagonises membrane localizaton | 42 | |
| Anillin | DID | cytokinesis, localization of mDia2 | 43 | |
| IQGAP | DID | phagocytosis, caveolae membrane insertion | 46 , 167 | |
| APC | DAD | synergistic actin polymerization | 51 - 53 , 64 | |
| INF2 | DAD | lamellipodia in podocyte | actin polymerization and SRF activation | 56 , 168 |
| Fli-I | DAD | promotes release of Dia autoinhition | 54 | |
| Hck | neutrophil chemotaxis | 148 | ||
| HDAC6 | MT deacetylation | 169 | ||
| PKD2 | N-terminal | spindle localization of PKD2 | 73 | |
| Exportin 6 | nuclear export | 170 | ||
| Importin-a | N-terminal | nuclear import | 171 | |
| Crm1 | C-terminal | nuclear export | 171 |
The FH1 domain has also been reported to bind to the intracellular part of the RAGE receptor protein (receptor for advanced glycation end products) and by this may mediate RAGE-dependent Rac1 and Cdc42 activation and cell migration.28 The molecular mechanism of Dia mediated RAGE signal transduction and its potential role on actin polymerization is unknown, however.
The FH2 domain, which forms a dimeric torus, is sufficient for actin nucleation. It can bind G-actin and the barbed end of filaments.29-31 As actin dimer and trimer formation is the kinetic barrier of actin polymerization, nucleation may be triggered by binding of actin mono-, di-, or trimers to the FH2 torus through weak actin binding sites.29 In addition to nucleation, Dia also elongates existing actin filaments, which depends on the G-actin binding protein Profilin.20,32 The FH2 torus remains bound to the barbed end of F-actin and may rotate during elongation.33 For elongation, one of the subunits of the dimeric torus loses its link to the barbed end, accepts a new actin monomer from a Profilin-actin complex, and adds it to the barbed end of the filament. In the next step, the other subunit of the FH2 torus dissociates from the barbed end and incorporates a new monomer. This stepwise mechanism is referred to as processive capping. A second, indirect mechanism contributes to elongation in vivo. The FH2 torus prevents binding of capping proteins to the barbed end, which counteracts elongation.
Ena/Vasp and Clip-170 have been described to physically interact with the FH2 domain of Dia.34-37 Although it has not been addressed, if and how binding of Ena/Vasp or Clip-170 interferes with nucleation and elongation and barbed end binding, it is attractive to speculate that binding to the FH2 domain may block sites important for the catalytic mechanism and suppress one or more of these activities. Future in vitro experiments, especially single molecule TIRF assays, will reveal such potential activities.
The RBD/FH3 domain is not a single entity in structural terms but includes a dimerization domain (DD), a coiled-coil domain (CC), and a domain with 4x Arm repeats.38-40 On the N-terminal side is the RBD that interacts with activated RhoGTPases (Fig. 1). As the autoregulatory domain at the C-terminal part (DAD) interacts with the 4x Arm domain and part of the RBD and thus prevents binding of activated RhoGTPases, this region is often refered to as Dia-inhibitory domain (DID).13,16,38,41 A number of structurally diverse proteins (Neurochondrin, Abi, Liprin, Anilin, IQGAP) bind to the FH3 domain.42-46 For none of these binding proteins a direct effect on actin polymerization has been reported yet. However, some of the FH3 interactors, Liprin, Anilin, and IQGAP, for example, seem to be involved in subcellular localization of Dia.42,43,46
In addition to these FH3 interactors, basic regions at the N-terminus or C-terminus of Dia are involved in controlling membrane association and binding of phosphoinositols (PIP). The N-terminal part contains a stretch of basic residues that is important for membrane association of Dia.47-49 In polarized Drosophila tracheal cells and Madin-Darby canine kidney (MDCK) cells, restriction of Dia to the apical domain is controlled by a combination of these mechanisms. On the one side, the RBD domain binds to apically activated and membrane attached Rho; on the other side, the basic N-terminal region of Dia binds to PI(4,5)P2, which is enriched in the apical membrane.47 The combination of these mechanisms not only mediates apical restriction but also controls activation of actin polymerization. Besides the release of DID-DAD autoinhibition by Rho-GTP binding, the interaction with PI(4,5)P2 may also contribute to activation. Such a dual mechanism is consistent with the previous report that addition of liposomes affects actin polymerization in a PIP2 concentration dependent manner.49
The DAD domain at the C-terminus tightly binds to the N-terminal DID domain to form a dormant Dia dimer.50 Only after opening of the intramolecular loop by binding of Rho GTPases to RBD, the FH2 torus becomes accessible to actin monomers and filaments, which allows nucleation and elongation. Similar to the disruption of the DID-DAD interaction by RhoGTPases, other proteins binding to the DAD domain may also release autoinhibition. APC (adenomatous polyposis coli) synergistically activates actin polymerization by binding to the DAD domain.51-53 Interestingly, APC and Dia together nucleate filaments but become separated upon elongation of the filament as demonstrated by single molecule TIRF microscopy. While Dia sits at the growing barbed end, APC remains associated with the pointed end.52 Flightless-1 (Fli-1), a member of the gelsolin family, is another example of an activating DAD interactor, which acts synergistically to Rho. As Rho-GTP only partially relieves autoinhibition, addition of Fli-1 to such an in vitro assay further activates Dia.54 Similar to proteins binding to the RBD or DAD, a synergistic activation may be achieved by phosphorylation of the serine or threonine residues within the DAD domain. Phosphorylation of mDia2 at 2 conserved residues (T1061 and S1070) by Rho dependent kinase (Rok) increases activation.55 DID and DAD domains of different members of the formin family may interact. Recent studies further showed that the DID domain of INF2 (inverted formin 2) binds to the DAD domain of mDia, allowing cross-regulation among formins. Such an interaction may be functionally relevant, as binding of INF2’s DID domain reduces the rate of actin polymerization by mDia1 in vitro.56
Physiological functions of Dia in single cells and in multicellular context
Essential role of Dia in cell cycle and cytokinesis
Dia was initially identified in screens for genes required for spermatogenesis in male flies.1,57,58 Counterparts of Dia in yeast, Bni1, Cdc12, and C. elegans, Cyk-1, were found because of their essential function in cytokinesis,18,59-63 demonstrating the conservation of Dia's function in cytokinesis.22,43,64-69 Dia is involved in setting up, positioning, and constriction of the contractile ring (Fig. 3). Loss of Dia function leads to failure of cytokinesis and results in multinuclear cells. In addition to cytokinesis, Dia is involved in diverse aspects of mitosis. Dia is required for proper segregation of centrosomes shortly before mitosis in Drosophila blastoderm embryos.70 Incomplete centrosome separation probably reflects an indirect function of Dia, as no obvious centrosomal localization of Dia has been reported yet. Dia has been implicated in the attachment of metaphase chromosomes to kinetochores, their alignment, as well as the orientation of the mitotic spindle and localization of spindle proteins.71-75 Dia may also be involved in linking cell growth with cell cycle progression at the G1/S transition.76,77 In the early Drosophila embryo, Dia is required for centrosome induced formation of pole cells, the germ cell precursors at the posterior pole of the embryo.69
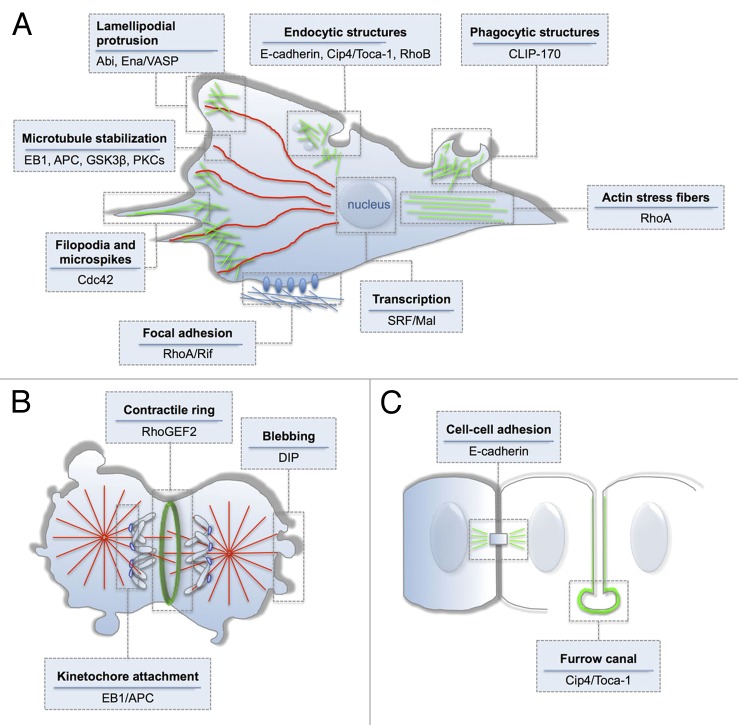
Figure 3. Dia dependent cellular structures and functions. Members of Diaphanous (Dia) protein family play important roles in (A) migrating, (B) in dividing, and (C) in the epithelial cells. Dia proteins mainly act on the assembly of actin filaments (marked in green) but also contribute to the regulation of microtubules dynamics (marked in red). Diverse cellular structures and functions are differentially regulated by a combinatorial use of numerous Dia binding partners (depicted in the boxes).
The role of Dia in regulating membrane protrusions and cell migration
Actin polymerization provides the mechanical force to drive membrane protrusions such as lamellipodia and filopodia. Formins, like Dia, nucleate linear actin filaments in filopodial protrusions (Fig. 3), whereas the Arp2/3 complex is thought to be the key nucleator generating a branched actin network during lamellipodia formation. Recent studies indicate that both protrusive structures require an intricate crosstalk between central actin nucleators. In the absence of the Arp2/3 complex or its major activator, the WAVE regulatory complex (WRC), mDia2-dependent filopodia are induced.78 By contrast, loss of mDia2 not only inhibits filopodia formation but also severely affects lamellipodia structure.79 All active mDia proteins (mDia1–3) are able to induce filopodia, although the underlying pathways seem to be different.80-82 An essential role of Dia in regulating lamellipodial-filopodial balance has been observed during Drosophila dorsal closure.83 Like mDia proteins, active Drosophila Dia localizes to filopodial tips and increases filopodial lifetime. In addition to such a direct function, Dia can also promote lamellipodia protrusions by recruiting Ena/VASP to the leading edge.83 Ena/VASP directly binds a central WRC subunit, Abi, the interactor of Abelson kinase (Abl),84 suggesting that filopodial assembly might originate from the lamellipodial network through the convergent-elongation mechanism as proposed previously.85 Supporting this notion, loss of function studies in different systems revealed conserved roles of Dia proteins in cell spreading and in cell migration including fibroblasts, epithelial, neuronal, and blood cells.79,83,86-88
The role of Dia in regulating endocytosis and endosome dynamics
Actin polymerization reshapes the plasma membrane but has also been proposed to drive membrane invaginations and propel endocytic vesicles during a variety of morphogenetic events. In the Drosophila embryo, Dia and non-muscle Myosin II (Myo II) control the initiation of E-cadherin endocytosis by regulating the recruitment of clathrin and the Adaptor Protein 2 (AP2).89 Thus, the function of Dia-dependent linear F-actin appears to be different from Arp2/3 induced branched actin filaments that promotes endocytosis. Unlike Dia, Arp2/3 mediated branched actin filament nucleation by WASP/WAVE proteins facilitates Dynamin-dependent vesicular scission.90,91 Recent live-imaging analyses of dia mutant embryos revealed an increased endocytic activity and thus suggests an inhibitory function of Dia on tubular membrane invaginations. This inhibitory role of Dia is based on an antagonistic interaction with the F-BAR protein Cip4, a known activator of the WASP/WAVE-Arp2/3 pathway.22,90-92 Since Cip4 inhibits actin nucleation by Dia in vitro, a model has been proposed in which Cip4 controls 2 different pools of actin filaments: activation of branched filaments by its WASP/WAVE interaction and suppression of linear filaments by inhibition of Dia.22 The strong co-localization of Dia and Cip4 at newly formed endocytic vesicles further suggests an additional role of Dia in vesicle trafficking.22 In mammals, a conserved function of Dia proteins in regulating endosome movement has been found. mDia1 and mDia2 are recruited to endosomes by activated RhoB.93-95 Endosomal F-actin induced by Dia associates with cortical actin stress fibers, which might control further transport of endosomes to microtubules.95 A similar function of mDia2 and Abl has recently been postulated for the formation of stress-fiber-linked caveolae.96
In addition to trafficking, Dia regulates the morphology of the Golgi apparatus.97 Activated Dia leads to dispersion of Golgi membrane stacks, which is based on repressed fusion of small Golgi stacks into larger compartments as well as increased formation of Rab6 positive transport vesicles. As Dia colocalized with the Rab6 vesicles, at least partially, their number may be directly controlled by Dia.97 Mitochondria are also affected by Dia. Activation of Dia leads to increased anchoring to actin filaments and loss of mobility of mitochondria, whereas RNAi mediated depletion of Dia elevated mobility.98
The role of Dia proteins on microtubule dynamics
Over the last decade, growing evidence has emerged that Dia not only acts on actin polymerization but also contributes to the regulation of microtubule dynamics.99 The first hints came from overexpression experiments with constitutively active mutant proteins lacking their autoregulatory domains. Expression of active mDia1 induces bipolar elongation of HeLa cells, in which microtubules are aligned in parallel with F-actin stress fibers.100 At the same time, Gregg Gunderson’s group observed that expression of active mDia2 is sufficient to generate and orient stable, detyrosinated microtubules.101 Subsequent work of the same group identified mDia2 in a complex with EB1 and APC, 2 important microtubule plus-end tracking proteins, suggesting a role for mDia2 in microtubule capping.102 The molecular complexity of mDia induced microtubule stabilization was further increased by the observation that mDia1 regulates the glycogen synthetase kinase-3β (GSK3β) through novel PKCs to promote microtubule stabilization.103
The molecular mechanism of microtubule stabilization by mDia proteins remained completely unclear for a long time. The first experimental evidence against an unspecific effect of mDia induced actin assembly on microtubule dynamics came from the finding that the microtubule stabilization activity can be separated from its actin nucleation activity.104 Actin nucleation-defective FH2 fragments of mDia1, mDia2, and mDia3 retain their ability to bind to microtubules, EB1, and APC, and more importantly, they are still capable to induce microtubule stabilization upon overexpression in serum-starved NIH3T3 cells.72,104 Thus, the FH2 domain seems to function in both actin nucleation and microtubule stabilization. Interestingly, re-expression of actin-nucleation-deficient mDia3 in mDia3 knockdown cells fully rescue the chromosome misalignment phenotype, suggesting that mDia3 directly acts on microtubules at the kinetochore.72,75 Since the activity of mDia2 for actin is in the nanomolar range, whereas that for microtubules is in the micromolar range, a competition model has been proposed, in which a redistribution of the mDia proteins to microtubules can only occur, if the affinity for microtubules is significantly increased.104 Supporting this idea, recent work from the Gundersen group identified an actin capping protein that promotes microtubule stabilization by antagonizing mDia1.105 Comparative in vitro studies with different recombinant formins further revealed a reciprocal inhibition between actin and microtubule dynamics.106 Microtubules strongly inhibit actin polymerization by mDia2, whereas actin monomers inhibit in turn microtubule binding/bundling by inverted formins such as INF2.106 Based on the observation that stable microtubules preferentially form at the leading edge of migrating cells,107 the current model proposes a sequential action of mDia proteins on actin filaments and microtubules. mDia proteins are released from actin filaments by competition through capping proteins to promote microtubule stabilization required for cell polarization. Despite increasing in vitro evidence, a physiological role of the microtubule stabilizing activity of Dia proteins has not yet been found in vivo. In Drosophila, Dia directly binds both fly APC’s (APC1 and APC2), but unlike in vertebrates, the APC-Dia complexes seem to affect actin directly rather than through an EB1-dependent effect on microtubules.51,64 However, known formin-dependent fundamental processes such as axonal outgrowth and growth cone motility, meiosis in mammalian oocytes or cytoplasmic streaming of Drosophila oocytes require a tightly regulated, spatiotemporal coordination between actin and microtubule dynamics. Thus, future studies will be required to further decipher the microtubule stabilizing activity of Dia proteins in the context of a living animal.
Roles of Dia in shaping tissues during development
Dia is required for embryonic development in multiple species. In zebrafish, Dia controls cell movements and convergent extension during gastrulation possibly as an effector of the wnt signaling pathway.108,109 In embryos depleted of Dia2, cells of the deep marginal layer, prechordal plate, and lateral epidermis lose protrusions and blebs. In Drosophila embryogenesis, formation of the first epithelial cell layer is controlled by Dia.69,110 During cellularization of dia mutant embryos, the array of invaginating furrows is disrupted and incomplete, and adjacent nuclei are frequently incorporated in the same cell. As the speed of invagination of formed furrows is not affected, Dia seems to stabilize newly formed furrows. In addition, the typical epithelial compartmentalization is impaired in dia mutants, as lateral and basal domains are not separated.22 In this process, Dia is controlled by RhoGEF2-Rho1 signaling and acts together with the non-receptor tyrosine kinase Abl and Ena (Enabled) in controlling actin filament formation in the apical microvillous structure of the plasma membrane.110,111 Later in embryonic development, Dia is involved in tissue morphogenesis. Invagination of the mesoderm, which relies on a stereotypic series of cell shape changes starting with apical constriction, is impaired in dia mutants, as the level of apical non-muscle myosin is reduced and the maturation of adherens junctions is disturbed.83 Specifically, Dia assembles F-actin filaments that suppress E-Cadherin localization in the medial region and connect contractile medial actomyosin filaments with adherens junctions.112 In dynamic junctions during cell intercalation, Dia promotes the turnover of E-Cadherin and thus contributes to establishing the anisotropy of actomyosin activity needed for directional intercalation. It was proposed that Dia controls formation of stable cortical actin patches, which lead to lateral clustering of E-Cadherin.89,113 Segmentation of Drosophila embryos becomes visible by segmental grooves, which are formed by infoldings of the epidermis. In this process, Dia may be involved in stabilization of adherens junctions but less so by triggering apical actomyosin contraction.114 Beside embryonic tissues, imaginal discs of Drosophila are an excellent system to analyze tissue remodelling and junction dynamics. It has been reported that Dia sustains apical tension during differentiation in the pupal eye disc.115
The function of Dia in formation, positioning, and maturation of cellular junctions has also been studied in cultured cells. Dia localizes to adherens junctions and is needed for maintenance and strengthening of adherens junctions downstream of Rho1 and counteracting Rho kinase.116-118 The requirement of Dia for adherens junctions may change during tumorigenesis. While Dia is involved in the assembly of tangential junctions in non-transformed cells, radial junctions are not Dia-dependent in transformed cells.116 Dia function may be coordinated by Arp2/3 through its direct interaction with Abi, a well-characterized WAVE/Arp2/3 activator.44 Studies of mice lacking mDia1 and mDia3 confirmed these results from cultured cells, showing that neuroepithelial cells have an attenuated apical actin belt and lost adherens junctions.119
Recent studies further highlighted the importance of apical targeting of Dia activity as a conserved feature of all epithelial cells forming different tubular organs in 3D cyst of cultured MDCK cells and in Drosophila trachea, salivary glands, hindgut, and Malpighian tubules.47,120 Remarkably, in the absence of apical actin polymerization, the apical-basal polarity of these tubular epithelial cells is not affected but secretion via the apical surface to the tube lumen is blocked.120 Apical secretion requires Myosin V (Myo V) motor protein, which transports secretory vesicles along polarized actin filaments nucleated by Dia.120
In addition to polarization, Dia is involved in the formation and the maturation of cell-cell and cell-matrix junctions. Drosophila neuromuscular junctions grow in a Dia-dependent manner after initial contact formation. Dia acts on the presynaptic side and is regulated by the receptor tyrosine phosphatase Dlar and the guanine nucleotide exchange factor Trio.121 Dia is also involved in the interaction and stimulation of dendritic cells by T cells as shown with bone-marrow derived dendritic cells from mDia1 deficient mice.122 Furthermore, Dia functions in focal adhesions and cell-matrix interactions. This function of Dia may be developmentally regulated and depends on the specific composition of FAs. During oogenesis in Drosophila, the follicle epithelium switches from a columnar to squamous morphology. This morphology change is associated with a switch in integrin subtypes. Analysis of mutant clones showed that integrin downregulated Dia and Profilin levels.123 Dia may be involved in maturation of focal adhesions as a mechano-sensor. The increased F-actin assembly and FA growth in focal adhesions induced by external force depends on mDia1.124 In addition, Dia may control assembly of the stress fibers that are linked to FA involving signaling by Rho or Rif GTPases.125-128
Dia-linking cytoskeleton with mechanics and transcription
Cells are able to respond to mechanical stimuli, such as stretching during cell migration by changes in transcription. The serum response factor (SRF) and its cofactor Mal constitute a pathway that mediates such a response and that is dependent on the pool of G-actin. SRF/Mal can be triggered by cytoplasmic and nuclear Dia in cultured cells.30,37,129-132 In response to forces, mDia1 promotes α-SMA (α smooth muscle actin) promoter activity as a result of the release of MRTF-A, a transcriptional co-activator of SMA from actin monomers. Conversely, force-induced α-SMA expression is blocked in mDia1 knockdown cells.133 Thus, differentiation of myofibroblasts depends on mDia1.133 The SRF/Mal pathway has been also investigated in its physiological context in collective migration of border cells in Drosophila oogenesis. Here, SRF/Mal provides a feedback mechanism for re-enforcing cytoskeletal strength. SRF/Mal signaling is essential for border cell migration, as mal mutant cells fail to migrate. Furthermore, accumulation of Mal in border cell nuclei is triggered by cell streching and activated Dia.134
Dia can sense and respond to mechanical stimuli.135 This may rely on indirect signaling mechanisms or availability of globular actin.136 In addition, the mechanism of progressive capping by the FH2 dimer appears to be inherently sensitive to forces in a piconewton range.32,137,138 This prediction has recently been tested experimentally with microfluidic devices, in which calibrated piconewton forces can be applied to actin filaments. The measurements showed that the elongation rate of polymerization by Dia increased by up to a factor of 2 when filaments were pulled.139 Such a force dependent activity may be a common feature of formins, as yeast Bni1 shows an elevated elongation rate after force application on filaments.140 Controlled mechanical deformations of the cell cortex induced processive F-actin assembly by Dia in a manner independent of Rho or Ca2+ ions but dependent on LIM kinase. In this system, the initial event that senses the mechanical changes may be an increased amount of globular actin monomers, which may in turn increase polymerization rate by Dia.136
Conversely to sensing, Dia may also directly control the mechanical properties of cells. The stiffness of cells is controlled by the actin cortex beneath the plasma membrane. Expression of activated Dia leads to an increased stiffness of cells141 and expression of Dia-interacting protein (DIP) induced mDia2 dependent blebbing of the plasma membrane,25 indicating a role of Dia in assembly of the actin cortex.
Functions in mammalian physiology
Most studies on Dia functions were focused on molecular and cellular aspects. Based on findings from human genetics and analyses in different genetic model organisms, unexpected specific functions in complex physiological processes have been identified. Very early on in the study of Dia, a link of Dia to deafness in humans was established. DFNA1, characterized by a fully penetrant sensorineural hearing loss and malfunction of hair cells in the inner ear, is linked to a mutation in Dia that leads to a small C-terminal truncation.142,143 A corresponding auditory dysfunction was observed in Drosophila dia mutants,144 indicating a conserved function. Conversely, hyperactivation of Dia/Dia3 in the auditory system also impairs hearing in Drosophila and mice and leads to auditory neuropathy (AUNA1) in humans.145,146 Another insight from human genetics was provided by the mapping of premature ovarian failure (POF) to the Dia3 gene.147 Although the consequences of the mutation in humans have not been reported yet, similarities to the function of Dia in the male and female germline of Drosophila1,69 may be expected.
In cells of the hematopoetic system, a number of specific functions of Dia have been revealed. Dia mediates adhesive and migratory behavior of dendritic cells and their interaction with T cells in the lymphnodes.122 Similarly, T cells and neutrophils deficient for mDia1 showed a poor adherence to the extracellular matrix and chemotactic migration behavior.25,148-150 These observations point to a role of Dia activation in T cell response.151
Dia is also important in the erythrocyte lineage. A unique feature of erythropoesis is the loss of the nucleus during maturation of erythroblasts. In a type of asymmetric cell division, the pyknotic nucleus moves to one “daughter” cell separated from the main, anuclear cell body by a contractile actin ring. Closure of the actin ring leads to extrusion of the nucleus. Dia2 and Rac GTPases control the formation of the contractile actin ring, while Dia depletion blocked enucleation.152 In platelets, Dia participates in thrombin-induced reorganization of the actin cytoskeleton.153,154 In macrophages, Dia is involved in phagocytosis. Dia together with formin-like protein 1 (FMNL1) controls the formation of Borrelia-induced pseudopods that capture and enwrap the pathogene,155 possibly by a mechanism that requires IQGAP and CLIP170 functions.39,46,156 A requirement of Dia has been shown during pathogen infection, such as Shigella. The virulence protein IpgB2 binds the Rho binding domain of Dia and is involved in protrusion formation and spread of bacteria within the host cell.157,158
Finally, a surprising link of Dia to cortisol hormones was identified in adrenocortical cells.159,160 In response to adrenocorticotropin (ACTH) steroids are synthesized with the reaction pathway partly located in the endoplasmic reticulum and mitochondria. As interference with Dia1 lead to decreased mitochondria mobility and decreased cortisol but increased adrenal androgene synthesis, Dia1 seems to be involved in interorganelle substrate transfer by dynamic trafficking of mitochondria.
Conclusions and Perspectives
In summary, Dia proteins control diverse fundamental biological processes that are mediated by combinatorial interactions with different binding partners. The nature of some interactions including well-known actin regulators such as Abi proteins or members of the Ena/VASP proteins is still unclear. Future genetic approaches including detailed structure-function analyses in the mutant background will significantly increase our understanding of these interactions in vivo. Additional quantitative multi-wavelength single-molecule imaging approaches and new super-resolution microscopy approaches will shed light on these interactions how and where these conserved regulatory networks act on cellular structures and cell dynamics. The use of advanced optogenetic tools for the manipulation of endogenous proteins in single cells will further complement our knowledge about these evolutionarily conserved modules in actin nucleation.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank C Klämbt for critical reading of the manuscript. This work was supported by the priority program “Actin nucleators” (SPP1464) from the Deutsche Forschungsgemeinschaft.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cib/article/27634
References
- 1.Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–77. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- 2.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–51. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schönichen A, Geyer M. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim Biophys Acta. 2010;1803:152–63. doi: 10.1016/j.bbamcr.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–53. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Rivero F, Muramoto T, Meyer AK, Urushihara H, Uyeda TQ, Kitayama C. A comparative sequence analysis reveals a common GBD/FH3-FH1-FH2-DAD architecture in formins from Dictyostelium, fungi and metazoa. BMC Genomics. 2005;6:28. doi: 10.1186/1471-2164-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalkia D, Nikolaidis N, Makalowski W, Klein J, Nei M. Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol Biol Evol. 2008;25:2717–33. doi: 10.1093/molbev/msn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grunt M, Zárský V, Cvrcková F. Roots of angiosperm formins: the evolutionary history of plant FH2 domain-containing proteins. BMC Evol Biol. 2008;8:115. doi: 10.1186/1471-2148-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–5. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 13.Lammers M, Meyer S, Kühlmann D, Wittinghofer A. Specificity of interactions between mDia isoforms and Rho proteins. J Biol Chem. 2008;283:35236–46. doi: 10.1074/jbc.M805634200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol Biol Evol. 2001;18:453–64. doi: 10.1093/oxfordjournals.molbev.a003824. [DOI] [PubMed] [Google Scholar]
- 15.Mazin PV, Gelfand MS, Mironov AA, Rakhmaninova AB, Rubinov AR, Russell RB, Kalinina OV. An automated stochastic approach to the identification of the protein specificity determinants and functional subfamilies. Algorithms Mol Biol. 2010;5:29. doi: 10.1186/1748-7188-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005;18:273–81. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Kursula P, Kursula I, Massimi M, Song YH, Downer J, Stanley WA, Witke W, Wilmanns M. High-resolution structural analysis of mammalian profilin 2a complex formation with two physiological ligands: the formin homology 1 domain of mDia1 and the proline-rich domain of VASP. J Mol Biol. 2008;375:270–90. doi: 10.1016/j.jmb.2007.10.050. [DOI] [PubMed] [Google Scholar]
- 18.Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–82. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–56. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–29. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 21.Kovar DR, Pollard TD. Progressing actin: Formin as a processive elongation machine. Nat Cell Biol. 2004;6:1158–9. doi: 10.1038/ncb1204-1158. [DOI] [PubMed] [Google Scholar]
- 22.Yan S, Lv Z, Winterhoff M, Wenzl C, Zobel T, Faix J, Bogdan S, Grosshans J. The F-BAR protein Cip4/Toca-1 antagonizes the formin Diaphanous in membrane stabilization and compartmentalization. J Cell Sci. 2013;126:1796–805. doi: 10.1242/jcs.118422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satoh S, Tominaga T. mDia-interacting protein acts downstream of Rho-mDia and modifies Src activation and stress fiber formation. J Biol Chem. 2001;276:39290–4. doi: 10.1074/jbc.M107026200. [DOI] [PubMed] [Google Scholar]
- 24.Fujiwara T, Mammoto A, Kim Y, Takai Y. Rho small G-protein-dependent binding of mDia to an Src homology 3 domain-containing IRSp53/BAIAP2. Biochem Biophys Res Commun. 2000;271:626–9. doi: 10.1006/bbrc.2000.2671. [DOI] [PubMed] [Google Scholar]
- 25.Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol. 2007;17:579–91. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Esue O, Harris ES, Higgs HN, Wirtz D. The filamentous actin cross-linking/bundling activity of mammalian formins. J Mol Biol. 2008;384:324–34. doi: 10.1016/j.jmb.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 27.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–92. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 28.Hudson BI, Kalea AZ, Del Mar Arriero M, Harja E, Boulanger E, D’Agati V, Schmidt AM. Interaction of the RAGE cytoplasmic domain with diaphanous-1 is required for ligand-stimulated cellular migration through activation of Rac1 and Cdc42. J Biol Chem. 2008;283:34457–68. doi: 10.1074/jbc.M801465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kupi T, Gróf P, Nyitrai M, Belágyi J. Interaction of formin FH2 with skeletal muscle actin. EPR and DSC studies. Eur Biophys J. 2013;42:757–65. doi: 10.1007/s00249-013-0922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Copeland JW, Copeland SJ, Treisman R. Homo-oligomerization is essential for F-actin assembly by the formin family FH2 domain. J Biol Chem. 2004;279:50250–6. doi: 10.1074/jbc.M404429200. [DOI] [PubMed] [Google Scholar]
- 31.Shimada A, Nyitrai M, Vetter IR, Kühlmann D, Bugyi B, Narumiya S, Geeves MA, Wittinghofer A. The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol Cell. 2004;13:511–22. doi: 10.1016/S1097-2765(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 32.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004;101:14725–30. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno H, Higashida C, Yuan Y, Ishizaki T, Narumiya S, Watanabe N. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331:80–3. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- 34.Homem CC, Peifer M. Exploring the roles of diaphanous and enabled activity in shaping the balance between filopodia and lamellipodia. Mol Biol Cell. 2009;20:5138–55. doi: 10.1091/mbc.E09-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewkowicz E, Herit F, Le Clainche C, Bourdoncle P, Perez F, Niedergang F. The microtubule-binding protein CLIP-170 coordinates mDia1 and actin reorganization during CR3-mediated phagocytosis. J Cell Biol. 2008;183:1287–98. doi: 10.1083/jcb.200807023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schirenbeck A, Arasada R, Bretschneider T, Stradal TE, Schleicher M, Faix J. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc Natl Acad Sci U S A. 2006;103:7694–9. doi: 10.1073/pnas.0511243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grosse R, Copeland JW, Newsome TP, Way M, Treisman R. A role for VASP in RhoA-Diaphanous signalling to actin dynamics and SRF activity. EMBO J. 2003;22:3050–61. doi: 10.1093/emboj/cdg287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lammers M, Rose R, Scrima A, Wittinghofer A. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 2005;24:4176–87. doi: 10.1038/sj.emboj.7600879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li F, Higgs HN. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13:1335–40. doi: 10.1016/S0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- 40.Li F, Higgs HN. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem. 2005;280:6986–92. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- 41.Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435:513–8. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- 42.Sakamoto S, Ishizaki T, Okawa K, Watanabe S, Arakawa T, Watanabe N, Narumiya S. Liprin-α controls stress fiber formation by binding to mDia and regulating its membrane localization. J Cell Sci. 2012;125:108–20. doi: 10.1242/jcs.087411. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe S, Okawa K, Miki T, Sakamoto S, Morinaga T, Segawa K, Arakawa T, Kinoshita M, Ishizaki T, Narumiya S. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol Biol Cell. 2010;21:3193–204. doi: 10.1091/mbc.E10-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryu JR, Echarri A, Li R, Pendergast AM. Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol Cell Biol. 2009;29:1735–48. doi: 10.1128/MCB.01483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwaibold EM, Brandt DT. Identification of Neurochondrin as a new interaction partner of the FH3 domain of the Diaphanous-related formin Dia1. Biochem Biophys Res Commun. 2008;373:366–72. doi: 10.1016/j.bbrc.2008.06.042. [DOI] [PubMed] [Google Scholar]
- 46.Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rousso T, Shewan AM, Mostov KE, Schejter ED, Shilo BZ. Apical targeting of the formin Diaphanous in Drosophila tubular epithelia. Elife. 2013;2:e00666. doi: 10.7554/eLife.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorelik R, Yang C, Kameswaran V, Dominguez R, Svitkina T. Mechanisms of plasma membrane targeting of formin mDia2 through its amino terminal domains. Mol Biol Cell. 2011;22:189–201. doi: 10.1091/mbc.E10-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramalingam N, Zhao H, Breitsprecher D, Lappalainen P, Faix J, Schleicher M. Phospholipids regulate localization and activity of mDia1 formin. Eur J Cell Biol. 2010;89:723–32. doi: 10.1016/j.ejcb.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Maiti S, Michelot A, Gould C, Blanchoin L, Sokolova O, Goode BL. Structure and activity of full-length formin mDia1. Cytoskeleton (Hoboken) 2012;69:393–405. doi: 10.1002/cm.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaiswal R, Stepanik V, Rankova A, Molinar O, Goode BL, McCartney BM. Drosophila homologues of adenomatous polyposis coli (APC) and the formin diaphanous collaborate by a conserved mechanism to stimulate actin filament assembly. J Biol Chem. 2013;288:13897–905. doi: 10.1074/jbc.M113.462051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336:1164–8. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–96. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Higashi T, Ikeda T, Murakami T, Shirakawa R, Kawato M, Okawa K, Furuse M, Kimura T, Kita T, Horiuchi H. Flightless-I (Fli-I) regulates the actin assembly activity of diaphanous-related formins (DRFs) Daam1 and mDia1 in cooperation with active Rho GTPase. J Biol Chem. 2010;285:16231–8. doi: 10.1074/jbc.M109.079236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staus DP, Taylor JM, Mack CP. Enhancement of mDia2 activity by Rho-kinase-dependent phosphorylation of the diaphanous autoregulatory domain. Biochem J. 2011;439:57–65. doi: 10.1042/BJ20101700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun H, Schlondorff J, Higgs HN, Pollak MR. Inverted formin 2 regulates actin dynamics by antagonizing Rho/diaphanous-related formin signaling. J Am Soc Nephrol. 2013;24:917–29. doi: 10.1681/ASN.2012080834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giansanti MG, Bonaccorsi S, Gatti M. The role of anillin in meiotic cytokinesis of Drosophila males. J Cell Sci. 1999;112:2323–34. doi: 10.1242/jcs.112.14.2323. [DOI] [PubMed] [Google Scholar]
- 58.Gönczy P, DiNardo S. The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development. 1996;122:2437–47. doi: 10.1242/dev.122.8.2437. [DOI] [PubMed] [Google Scholar]
- 59.Ucar H, Tachibana K, Kishimoto T. The Mos-MAPK pathway regulates Diaphanous-related formin activity to drive cleavage furrow closure during polar body extrusion in starfish oocytes. J Cell Sci. 2013;126:5153–65. doi: 10.1242/jcs.130476. [DOI] [PubMed] [Google Scholar]
- 60.Piekny AJ, Mains PE. Rho-binding kinase (LET-502) and myosin phosphatase (MEL-11) regulate cytokinesis in the early Caenorhabditis elegans embryo. J Cell Sci. 2002;115:2271–82. doi: 10.1242/jcs.115.11.2271. [DOI] [PubMed] [Google Scholar]
- 61.Severson AF, Baillie DL, Bowerman B. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol. 2002;12:2066–75. doi: 10.1016/S0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- 62.Swan KA, Severson AF, Carter JC, Martin PR, Schnabel H, Schnabel R, Bowerman B. cyk-1: a C. elegans FH gene required for a late step in embryonic cytokinesis. J Cell Sci. 1998;111:2017–27. doi: 10.1242/jcs.111.14.2017. [DOI] [PubMed] [Google Scholar]
- 63.Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, et al. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–8. [PMC free article] [PubMed] [Google Scholar]
- 64.Webb RL, Zhou MN, McCartney BM. A novel role for an APC2-Diaphanous complex in regulating actin organization in Drosophila. Development. 2009;136:1283–93. doi: 10.1242/dev.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–38. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zavortink M, Contreras N, Addy T, Bejsovec A, Saint R. Tum/RacGAP50C provides a critical link between anaphase microtubules and the assembly of the contractile ring in Drosophila melanogaster. J Cell Sci. 2005;118:5381–92. doi: 10.1242/jcs.02652. [DOI] [PubMed] [Google Scholar]
- 67.Dean SO, Rogers SL, Stuurman N, Vale RD, Spudich JA. Distinct pathways control recruitment and maintenance of myosin II at the cleavage furrow during cytokinesis. Proc Natl Acad Sci U S A. 2005;102:13473–8. doi: 10.1073/pnas.0506810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kato T, Watanabe N, Morishima Y, Fujita A, Ishizaki T, Narumiya S. Localization of a mammalian homolog of diaphanous, mDia1, to the mitotic spindle in HeLa cells. J Cell Sci. 2001;114:775–84. doi: 10.1242/jcs.114.4.775. [DOI] [PubMed] [Google Scholar]
- 69.Afshar K, Stuart B, Wasserman SA. Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development. 2000;127:1887–97. doi: 10.1242/dev.127.9.1887. [DOI] [PubMed] [Google Scholar]
- 70.Cao J, Crest J, Fasulo B, Sullivan W. Cortical actin dynamics facilitate early-stage centrosome separation. Curr Biol. 2010;20:770–6. doi: 10.1016/j.cub.2010.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnston CA, Manning L, Lu MS, Golub O, Doe CQ, Prehoda KE. Formin-mediated actin polymerization cooperates with Mushroom body defect (Mud)-Dynein during Frizzled-Dishevelled spindle orientation. J Cell Sci. 2013;126:4436–44. doi: 10.1242/jcs.129544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng L, Zhang J, Ahmad S, Rozier L, Yu H, Deng H, Mao Y. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev Cell. 2011;20:342–52. doi: 10.1016/j.devcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rundle DR, Gorbsky G, Tsiokas L. PKD2 interacts and co-localizes with mDia1 to mitotic spindles of dividing cells: role of mDia1 IN PKD2 localization to mitotic spindles. J Biol Chem. 2004;279:29728–39. doi: 10.1074/jbc.M400544200. [DOI] [PubMed] [Google Scholar]
- 74.Narumiya S, Oceguera-Yanez F, Yasuda S. A new look at Rho GTPases in cell cycle: role in kinetochore-microtubule attachment. Cell Cycle. 2004;3:855–7. doi: 10.4161/cc.3.7.990. [DOI] [PubMed] [Google Scholar]
- 75.Yasuda S, Oceguera-Yanez F, Kato T, Okamoto M, Yonemura S, Terada Y, Ishizaki T, Narumiya S. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature. 2004;428:767–71. doi: 10.1038/nature02452. [DOI] [PubMed] [Google Scholar]
- 76.Kyrkou A, Soufi M, Bahtz R, Ferguson C, Bai M, Parton RG, Hoffmann I, Zerial M, Fotsis T, Murphy C. RhoD participates in the regulation of cell-cycle progression and centrosome duplication. Oncogene. 2013;32:1831–42. doi: 10.1038/onc.2012.195. [DOI] [PubMed] [Google Scholar]
- 77.Mammoto A, Huang S, Moore K, Oh P, Ingber DE. Role of RhoA, mDia, and ROCK in cell shape-dependent control of the Skp2-p27kip1 pathway and the G1/S transition. J Biol Chem. 2004;279:26323–30. doi: 10.1074/jbc.M402725200. [DOI] [PubMed] [Google Scholar]
- 78.Beli P, Mascheroni D, Xu D, Innocenti M. WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat Cell Biol. 2008;10:849–57. doi: 10.1038/ncb1745. [DOI] [PubMed] [Google Scholar]
- 79.Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goh WI, Ahmed S. mDia1-3 in mammalian filopodia. Commun Integr Biol. 2012;5:340–4. doi: 10.4161/cib.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goh WI, Lim KB, Sudhaharan T, Sem KP, Bu W, Chou AM, Ahmed S. mDia1 and WAVE2 proteins interact directly with IRSp53 in filopodia and are involved in filopodium formation. J Biol Chem. 2012;287:4702–14. doi: 10.1074/jbc.M111.305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mellor H. The role of formins in filopodia formation. Biochim Biophys Acta. 2010;1803:191–200. doi: 10.1016/j.bbamcr.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 83.Homem CC, Peifer M. Diaphanous regulates myosin and adherens junctions to control cell contractility and protrusive behavior during morphogenesis. Development. 2008;135:1005–18. doi: 10.1242/dev.016337. [DOI] [PubMed] [Google Scholar]
- 84.Maruoka M, Sato M, Yuan Y, Ichiba M, Fujii R, Ogawa T, Ishida-Kitagawa N, Takeya T, Watanabe N. Abl-1-bridged tyrosine phosphorylation of VASP by Abelson kinase impairs association of VASP to focal adhesions and regulates leukaemic cell adhesion. Biochem J. 2012;441:889–99. doi: 10.1042/BJ20110951. [DOI] [PubMed] [Google Scholar]
- 85.Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–21. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shinohara R, Thumkeo D, Kamijo H, Kaneko N, Sawamoto K, Watanabe K, Takebayashi H, Kiyonari H, Ishizaki T, Furuyashiki T, et al. A role for mDia, a Rho-regulated actin nucleator, in tangential migration of interneuron precursors. Nat Neurosci. 2012;15:373–80, S1-2. doi: 10.1038/nn.3020. [DOI] [PubMed] [Google Scholar]
- 87.Goulimari P, Kitzing TM, Knieling H, Brandt DT, Offermanns S, Grosse R. Galpha12/13 is essential for directed cell migration and localized Rho-Dia1 function. J Biol Chem. 2005;280:42242–51. doi: 10.1074/jbc.M508690200. [DOI] [PubMed] [Google Scholar]
- 88.Gupton SL, Gertler FB. Filopodia: the fingers that do the walking. Sci STKE. 2007;2007:re5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- 89.Levayer R, Pelissier-Monier A, Lecuit T. Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nat Cell Biol. 2011;13:529–40. doi: 10.1038/ncb2224. [DOI] [PubMed] [Google Scholar]
- 90.Fricke R, Gohl C, Dharmalingam E, Grevelhörster A, Zahedi B, Harden N, Kessels M, Qualmann B, Bogdan S. Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE. Curr Biol. 2009;19:1429–37. doi: 10.1016/j.cub.2009.07.058. [DOI] [PubMed] [Google Scholar]
- 91.Leibfried A, Fricke R, Morgan MJ, Bogdan S, Bellaiche Y. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr Biol. 2008;18:1639–48. doi: 10.1016/j.cub.2008.09.063. [DOI] [PubMed] [Google Scholar]
- 92.Fricke R, Gohl C, Bogdan S. The F-BAR protein family Actin’ on the membrane. Commun Integr Biol. 2010;3:89–94. doi: 10.4161/cib.3.2.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wallar BJ, Deward AD, Resau JH, Alberts AS. RhoB and the mammalian Diaphanous-related formin mDia2 in endosome trafficking. Exp Cell Res. 2007;313:560–71. doi: 10.1016/j.yexcr.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 94.Wallar BJ, Stropich BN, Schoenherr JA, Holman HA, Kitchen SM, Alberts AS. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J Biol Chem. 2006;281:4300–7. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- 95.Fernandez-Borja M, Janssen L, Verwoerd D, Hordijk P, Neefjes J. RhoB regulates endosome transport by promoting actin assembly on endosomal membranes through Dia1. J Cell Sci. 2005;118:2661–70. doi: 10.1242/jcs.02384. [DOI] [PubMed] [Google Scholar]
- 96.Echarri A, Muriel O, Pavón DM, Azegrouz H, Escolar F, Terrón MC, Sanchez-Cabo F, Martínez F, Montoya MC, Llorca O, et al. Caveolar domain organization and trafficking is regulated by Abl kinases and mDia1. J Cell Sci. 2012;125:3097–113. doi: 10.1242/jcs.090134. [DOI] [PubMed] [Google Scholar]
- 97.Zilberman Y, Alieva NO, Miserey-Lenkei S, Lichtenstein A, Kam Z, Sabanay H, Bershadsky A. Involvement of the Rho-mDia1 pathway in the regulation of Golgi complex architecture and dynamics. Mol Biol Cell. 2011;22:2900–11. doi: 10.1091/mbc.E11-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Minin AA, Kulik AV, Gyoeva FK, Li Y, Goshima G, Gelfand VI. Regulation of mitochondria distribution by RhoA and formins. J Cell Sci. 2006;119:659–70. doi: 10.1242/jcs.02762. [DOI] [PubMed] [Google Scholar]
- 99.DeWard AD, Alberts AS. Microtubule stabilization: formins assert their independence. Curr Biol. 2008;18:R605–8. doi: 10.1016/j.cub.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 100.Ishizaki T, Morishima Y, Okamoto M, Furuyashiki T, Kato T, Narumiya S. Coordination of microtubules and the actin cytoskeleton by the Rho effector mDia1. Nat Cell Biol. 2001;3:8–14. doi: 10.1038/35050598. [DOI] [PubMed] [Google Scholar]
- 101.Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol. 2001;3:723–9. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- 102.Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–30. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 103.Eng CH, Huckaba TM, Gundersen GG. The formin mDia regulates GSK3beta through novel PKCs to promote microtubule stabilization but not MTOC reorientation in migrating fibroblasts. Mol Biol Cell. 2006;17:5004–16. doi: 10.1091/mbc.E05-10-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bartolini F, Moseley JB, Schmoranzer J, Cassimeris L, Goode BL, Gundersen GG. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol. 2008;181:523–36. doi: 10.1083/jcb.200709029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bartolini F, Ramalingam N, Gundersen GG. Actin-capping protein promotes microtubule stability by antagonizing the actin activity of mDia1. Mol Biol Cell. 2012;23:4032–40. doi: 10.1091/mbc.E12-05-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gaillard J, Ramabhadran V, Neumanne E, Gurel P, Blanchoin L, Vantard M, Higgs HN. Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol Biol Cell. 2011;22:4575–87. doi: 10.1091/mbc.E11-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gundersen GG, Bulinski JC. Selective stabilization of microtubules oriented toward the direction of cell migration. Proc Natl Acad Sci U S A. 1988;85:5946–50. doi: 10.1073/pnas.85.16.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lai SL, Chan TH, Lin MJ, Huang WP, Lou SW, Lee SJ. Diaphanous-related formin 2 and profilin I are required for gastrulation cell movements. PLoS One. 2008;3:e3439. doi: 10.1371/journal.pone.0003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhu S, Liu L, Korzh V, Gong Z, Low BC. RhoA acts downstream of Wnt5 and Wnt11 to regulate convergence and extension movements by involving effectors Rho kinase and Diaphanous: use of zebrafish as an in vivo model for GTPase signaling. Cell Signal. 2006;18:359–72. doi: 10.1016/j.cellsig.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 110.Grosshans J, Wenzl C, Herz HM, Bartoszewski S, Schnorrer F, Vogt N, Schwarz H, Müller HA. RhoGEF2 and the formin Dia control the formation of the furrow canal by directed actin assembly during Drosophila cellularisation. Development. 2005;132:1009–20. doi: 10.1242/dev.01669. [DOI] [PubMed] [Google Scholar]
- 111.Grevengoed EE, Fox DT, Gates J, Peifer M. Balancing different types of actin polymerization at distinct sites: roles for Abelson kinase and Enabled. J Cell Biol. 2003;163:1267–79. doi: 10.1083/jcb.200307026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mason FM, Tworoger M, Martin AC. Apical domain polarization localizes actin-myosin activity to drive ratchet-like apical constriction. Nat Cell Biol. 2013;15:926–36. doi: 10.1038/ncb2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cavey M, Rauzi M, Lenne PF, Lecuit T. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 2008;453:751–6. doi: 10.1038/nature06953. [DOI] [PubMed] [Google Scholar]
- 114.Mulinari S, Barmchi MP, Häcker U. DRhoGEF2 and diaphanous regulate contractile force during segmental groove morphogenesis in the Drosophila embryo. Mol Biol Cell. 2008;19:1883–92. doi: 10.1091/mbc.E07-12-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Warner SJ, Longmore GD. Distinct functions for Rho1 in maintaining adherens junctions and apical tension in remodeling epithelia. J Cell Biol. 2009;185:1111–25. doi: 10.1083/jcb.200901029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ayollo DV, Zhitnyak IY, Vasiliev JM, Gloushankova NA. Rearrangements of the actin cytoskeleton and E-cadherin-based adherens junctions caused by neoplasic transformation change cell-cell interactions. PLoS One. 2009;4:e8027. doi: 10.1371/journal.pone.0008027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sahai E, Marshall CJ. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat Cell Biol. 2002;4:408–15. doi: 10.1038/ncb796. [DOI] [PubMed] [Google Scholar]
- 118.Nakano K, Takaishi K, Kodama A, Mammoto A, Shiozaki H, Monden M, Takai Y. Distinct actions and cooperative roles of ROCK and mDia in Rho small G protein-induced reorganization of the actin cytoskeleton in Madin-Darby canine kidney cells. Mol Biol Cell. 1999;10:2481–91. doi: 10.1091/mbc.10.8.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Thumkeo D, Shinohara R, Watanabe K, Takebayashi H, Toyoda Y, Tohyama K, Ishizaki T, Furuyashiki T, Narumiya S. Deficiency of mDia, an actin nucleator, disrupts integrity of neuroepithelium and causes periventricular dysplasia. PLoS One. 2011;6:e25465. doi: 10.1371/journal.pone.0025465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Massarwa R, Schejter ED, Shilo BZ. Apical secretion in epithelial tubes of the Drosophila embryo is directed by the Formin-family protein Diaphanous. Dev Cell. 2009;16:877–88. doi: 10.1016/j.devcel.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 121.Pawson C, Eaton BA, Davis GW. Formin-dependent synaptic growth: evidence that Dlar signals via Diaphanous to modulate synaptic actin and dynamic pioneer microtubules. J Neurosci. 2008;28:11111–23. doi: 10.1523/JNEUROSCI.0833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tanizaki H, Egawa G, Inaba K, Honda T, Nakajima S, Moniaga CS, Otsuka A, Ishizaki T, Tomura M, Watanabe T, et al. Rho-mDia1 pathway is required for adhesion, migration, and T-cell stimulation in dendritic cells. Blood. 2010;116:5875–84. doi: 10.1182/blood-2010-01-264150. [DOI] [PubMed] [Google Scholar]
- 123.Delon I, Brown NH. The integrin adhesion complex changes its composition and function during morphogenesis of an epithelium. J Cell Sci. 2009;122:4363–74. doi: 10.1242/jcs.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bershadsky AD, Ballestrem C, Carramusa L, Zilberman Y, Gilquin B, Khochbin S, Alexandrova AY, Verkhovsky AB, Shemesh T, Kozlov MM. Assembly and mechanosensory function of focal adhesions: experiments and models. Eur J Cell Biol. 2006;85:165–73. doi: 10.1016/j.ejcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 125.Rao MV, Chu PH, Hahn KM, Zaidel-Bar R. An optogenetic tool for the activation of endogenous diaphanous-related formins induces thickening of stress fibers without an increase in contractility. Cytoskeleton (Hoboken) 2013;70:394–407. doi: 10.1002/cm.21115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fan L, Pellegrin S, Scott A, Mellor H. The small GTPase Rif is an alternative trigger for the formation of actin stress fibers in epithelial cells. J Cell Sci. 2010;123:1247–52. doi: 10.1242/jcs.061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–94. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Anderson S, DiCesare L, Tan I, Leung T, SundarRaj N. Rho-mediated assembly of stress fibers is differentially regulated in corneal fibroblasts and myofibroblasts. Exp Cell Res. 2004;298:574–83. doi: 10.1016/j.yexcr.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 129.Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340:864–7. doi: 10.1126/science.1235038. [DOI] [PubMed] [Google Scholar]
- 130.Copeland JW, Treisman R. The diaphanous-related formin mDia1 controls serum response factor activity through its effects on actin polymerization. Mol Biol Cell. 2002;13:4088–99. doi: 10.1091/mbc.02-06-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Geneste O, Copeland JW, Treisman R. LIM kinase and Diaphanous cooperate to regulate serum response factor and actin dynamics. J Cell Biol. 2002;157:831–8. doi: 10.1083/jcb.200203126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 2000;5:13–25. doi: 10.1016/S1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- 133.Chan MW, Chaudary F, Lee W, Copeland JW, McCulloch CA. Force-induced myofibroblast differentiation through collagen receptors is dependent on mammalian diaphanous (mDia) J Biol Chem. 2010;285:9273–81. doi: 10.1074/jbc.M109.075218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Somogyi K, Rørth P. Evidence for tension-based regulation of Drosophila MAL and SRF during invasive cell migration. Dev Cell. 2004;7:85–93. doi: 10.1016/j.devcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 135.Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153:1175–86. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Higashida C, Kiuchi T, Akiba Y, Mizuno H, Maruoka M, Narumiya S, Mizuno K, Watanabe N. F- and G-actin homeostasis regulates mechanosensitive actin nucleation by formins. Nat Cell Biol. 2013;15:395–405. doi: 10.1038/ncb2693. [DOI] [PubMed] [Google Scholar]
- 137.Shemesh T, Geiger B, Bershadsky AD, Kozlov MM. Focal adhesions as mechanosensors: a physical mechanism. Proc Natl Acad Sci U S A. 2005;102:12383–8. doi: 10.1073/pnas.0500254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kozlov MM, Bershadsky AD. Processive capping by formin suggests a force-driven mechanism of actin polymerization. J Cell Biol. 2004;167:1011–7. doi: 10.1083/jcb.200410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jégou A, Carlier MF, Romet-Lemonne G. Formin mDia1 senses and generates mechanical forces on actin filaments. Nat Commun. 2013;4:1883. doi: 10.1038/ncomms2888. [DOI] [PubMed] [Google Scholar]
- 140.Courtemanche N, Lee JY, Pollard TD, Greene EC. Tension modulates actin filament polymerization mediated by formin and profilin. Proc Natl Acad Sci U S A. 2013;110:9752–7. doi: 10.1073/pnas.1308257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tamura K, Mizutani T, Haga H, Kawabata K. Nano-mechanical properties of living cells expressing constitutively active RhoA effectors. Biochem Biophys Res Commun. 2010;403:363–7. doi: 10.1016/j.bbrc.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 142.Lalwani AK, Jackler RK, Sweetow RW, Lynch ED, Raventós H, Morrow J, King MC, León PE. Further characterization of the DFNA1 audiovestibular phenotype. Arch Otolaryngol Head Neck Surg. 1998;124:699–702. doi: 10.1001/archotol.124.6.699. [DOI] [PubMed] [Google Scholar]
- 143.Lynch ED, Lee MK, Morrow JE, Welcsh PL, León PE, King MC. Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science. 1997;278:1315–8. doi: 10.1126/science.278.5341.1315. [DOI] [PubMed] [Google Scholar]
- 144.Cosetti M, Culang D, Kotla S, O’Brien P, Eberl DF, Hannan F. Unique transgenic animal model for hereditary hearing loss. Ann Otol Rhinol Laryngol. 2008;117:827–33. doi: 10.1177/000348940811701106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schoen CJ, Burmeister M, Lesperance MM. Diaphanous homolog 3 (Diap3) overexpression causes progressive hearing loss and inner hair cell defects in a transgenic mouse model of human deafness. PLoS One. 2013;8:e56520. doi: 10.1371/journal.pone.0056520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schoen CJ, Emery SB, Thorne MC, Ammana HR, Sliwerska E, Arnett J, Hortsch M, Hannan F, Burmeister M, Lesperance MM. Increased activity of Diaphanous homolog 3 (DIAPH3)/diaphanous causes hearing defects in humans with auditory neuropathy and in Drosophila. Proc Natl Acad Sci U S A. 2010;107:13396–401. doi: 10.1073/pnas.1003027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bione S, Sala C, Manzini C, Arrigo G, Zuffardi O, Banfi S, Borsani G, Jonveaux P, Philippe C, Zuccotti M, et al. A human homologue of the Drosophila melanogaster diaphanous gene is disrupted in a patient with premature ovarian failure: evidence for conserved function in oogenesis and implications for human sterility. Am J Hum Genet. 1998;62:533–41. doi: 10.1086/301761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shi Y, Dong B, Miliotis H, Liu J, Alberts AS, Zhang J, Siminovitch KA. Src kinase Hck association with the WASp and mDia1 cytoskeletal regulators promotes chemoattractant-induced Hck membrane targeting and activation in neutrophils. Biochem Cell Biol. 2009;87:207–16. doi: 10.1139/O08-130. [DOI] [PubMed] [Google Scholar]
- 149.Shi Y, Zhang J, Mullin M, Dong B, Alberts AS, Siminovitch KA. The mDial formin is required for neutrophil polarization, migration, and activation of the LARG/RhoA/ROCK signaling axis during chemotaxis. J Immunol. 2009;182:3837–45. doi: 10.4049/jimmunol.0803838. [DOI] [PubMed] [Google Scholar]
- 150.Sakata D, Taniguchi H, Yasuda S, Adachi-Morishima A, Hamazaki Y, Nakayama R, Miki T, Minato N, Narumiya S. Impaired T lymphocyte trafficking in mice deficient in an actin-nucleating protein, mDia1. J Exp Med. 2007;204:2031–8. doi: 10.1084/jem.20062647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vicente-Manzanares M, Rey M, Pérez-Martínez M, Yáñez-Mó M, Sancho D, Cabrero JR, Barreiro O, de la Fuente H, Itoh K, Sánchez-Madrid F. The RhoA effector mDia is induced during T cell activation and regulates actin polymerization and cell migration in T lymphocytes. J Immunol. 2003;171:1023–34. doi: 10.4049/jimmunol.171.2.1023. [DOI] [PubMed] [Google Scholar]
- 152.Ji P, Jayapal SR, Lodish HF. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat Cell Biol. 2008;10:314–21. doi: 10.1038/ncb1693. [DOI] [PubMed] [Google Scholar]
- 153.Gao GX, Dong HJ, Gu HT, Gao Y, Pan YZ, Wang YW, Yang Y, Chen XQ. [PI3-kinase mediates thrombin-induced platelet aggregation through mDia1 pathway.] Zhonghua Xue Ye Xue Za Zhi. 2010;31:176–80. [PubMed] [Google Scholar]
- 154.Higashi T, Ikeda T, Shirakawa R, Kondo H, Kawato M, Horiguchi M, Okuda T, Okawa K, Fukai S, Nureki O, et al. Biochemical characterization of the Rho GTPase-regulated actin assembly by diaphanous-related formins, mDia1 and Daam1, in platelets. J Biol Chem. 2008;283:8746–55. doi: 10.1074/jbc.M707839200. [DOI] [PubMed] [Google Scholar]
- 155.Naj X, Hoffmann AK, Himmel M, Linder S. The formins FMNL1 and mDia1 regulate coiling phagocytosis of Borrelia burgdorferi by primary human macrophages. Infect Immun. 2013;81:1683–95. doi: 10.1128/IAI.01411-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Colucci-Guyon E, Niedergang F, Wallar BJ, Peng J, Alberts AS, Chavrier P. A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr Biol. 2005;15:2007–12. doi: 10.1016/j.cub.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 157.Heindl JE, Saran I, Yi CR, Lesser CF, Goldberg MB. Requirement for formin-induced actin polymerization during spread of Shigella flexneri. Infect Immun. 2010;78:193–203. doi: 10.1128/IAI.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–45. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 159.Li D, Dammer EB, Lucki NC, Sewer MB. cAMP-stimulated phosphorylation of diaphanous 1 regulates protein stability and interaction with binding partners in adrenocortical cells. Mol Biol Cell. 2013;24:848–57. doi: 10.1091/mbc.E12-08-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li D, Sewer MB. RhoA and DIAPH1 mediate adrenocorticotropin-stimulated cortisol biosynthesis by regulating mitochondrial trafficking. Endocrinology. 2010;151:4313–23. doi: 10.1210/en.2010-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tsujita K, Itoh T, Kondo A, Oyama M, Kozuka-Hata H, Irino Y, Hasegawa J, Takenawa T. Proteome of acidic phospholipid-binding proteins: spatial and temporal regulation of Coronin 1A by phosphoinositides. J Biol Chem. 2010;285:6781–9. doi: 10.1074/jbc.M109.057018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, Sudhaharan T, Ahmed S. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem. 2008;283:20454–72. doi: 10.1074/jbc.M710185200. [DOI] [PubMed] [Google Scholar]
- 163.Meng W, Numazaki M, Takeuchi K, Uchibori Y, Ando-Akatsuka Y, Tominaga M, Tominaga T. DIP (mDia interacting protein) is a key molecule regulating Rho and Rac in a Src-dependent manner. EMBO J. 2004;23:760–71. doi: 10.1038/sj.emboj.7600095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fukumi-Tominaga T, Mori Y, Matsuura A, Kaneko K, Matsui M, Ogata M, Tominaga M. DIP/WISH-deficient mice reveal Dia- and N-WASP-interacting protein as a regulator of cytoskeletal dynamics in embryonic fibroblasts. Genes Cells. 2009;14:1197–207. doi: 10.1111/j.1365-2443.2009.01345.x. [DOI] [PubMed] [Google Scholar]
- 165.Wyse MM, Lei J, Nestor-Kalinoski AL, Eisenmann KM. Dia-interacting protein (DIP) imposes migratory plasticity in mDia2-dependent tumor cells in three-dimensional matrices. PLoS One. 2012;7:e45085. doi: 10.1371/journal.pone.0045085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Touré F, Fritz G, Li Q, Rai V, Daffu G, Zou YS, Rosario R, Ramasamy R, Alberts AS, Yan SF, et al. Formin mDia1 mediates vascular remodeling via integration of oxidative and signal transduction pathways. Circ Res. 2012;110:1279–93. doi: 10.1161/CIRCRESAHA.111.262519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wickström SA, Lange A, Hess MW, Polleux J, Spatz JP, Krüger M, Pfaller K, Lambacher A, Bloch W, Mann M, et al. Integrin-linked kinase controls microtubule dynamics required for plasma membrane targeting of caveolae. Dev Cell. 2010;19:574–88. doi: 10.1016/j.devcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sun H, Schlondorff JS, Brown EJ, Higgs HN, Pollak MR. Rho activation of mDia formins is modulated by an interaction with inverted formin 2 (INF2) Proc Natl Acad Sci U S A. 2011;108:2933–8. doi: 10.1073/pnas.1017010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Destaing O, Saltel F, Gilquin B, Chabadel A, Khochbin S, Ory S, Jurdic P. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci. 2005;118:2901–11. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- 170.Stüven T, Hartmann E, Görlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 2003;22:5928–40. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Miki T, Okawa K, Sekimoto T, Yoneda Y, Watanabe S, Ishizaki T, Narumiya S. mDia2 shuttles between the nucleus and the cytoplasm through the importin-alpha/beta- and CRM1-mediated nuclear transport mechanism. J Biol Chem. 2009;284:5753–62. doi: 10.1074/jbc.M806191200. [DOI] [PubMed] [Google Scholar]
- 172.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 174.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–52. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.