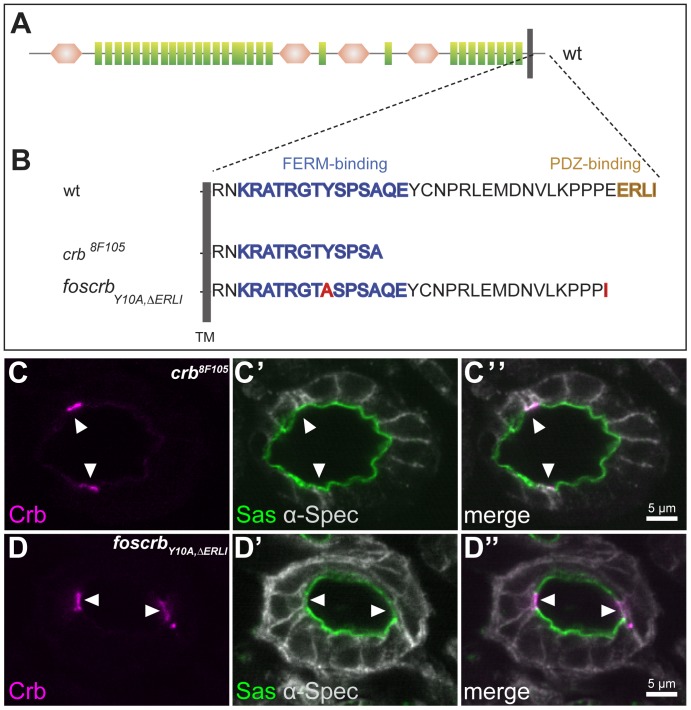
Figure 4. Localisation of Crb in BCs is independent from its known protein binding motifs.
(A) Schematic representation of the Crb protein and its variants used in this study. Green rectangles: EGF-like repeats, brown hexagons: laminin A G like domains, grey bar: transmembrane domain (TM). (B) Amino acid sequences of the cytoplasmic tails of wild-type and mutant Crb proteins used in this study. Blue: FERM domain-binding motif, brown: PDZ domain-binding motif. Red: point mutations. (C–D″) Confocal microscopy images of cross sections through the large intestine of stage 15 homozygous crb8F105 (C–C″) and foscrbY10A,ΔERLI (D–D″) embryos stained with anti-Crb (magenta), anti-α-Spectrin (grey) and anti-Sas (green). Crb is upregulated and apically localised in the BCs (white arrowheads), but is not detectable in the PCs.