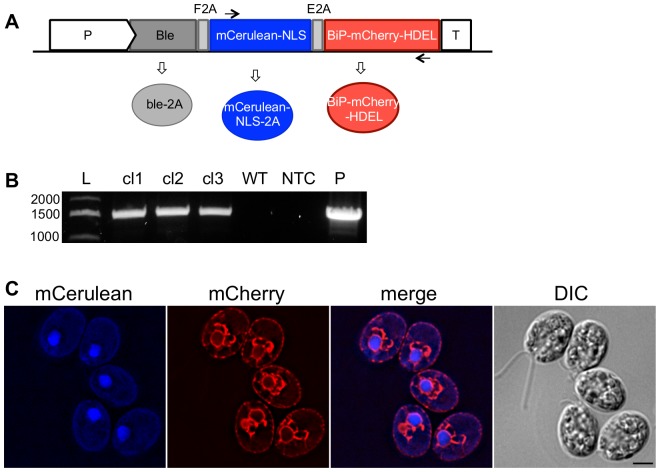
Figure 2. Gene stacking using a multi-cistronic transformation vector.
A. A schematic representation of the Chlamydomonas multi-cistronic expression vector. The expression of the cassette is under the control of the hsp70/rbcs2 promoter (P). Ble confers zeocin-resistance. mCerulean is targeted to the nucleus with the SV40-NLS. mCherry is targeted to the ER using the BiP ER-TS and the HDEL retention sequence. Two 2A self-cleaving sequences are fused between the three cistrons: F2A, from FMDV1; E2A, from equine rhinitis A virus. Black arrows represent the location of the oligonucleotides used in (B). Following co-translational processing of the 2A peptides, three distinct proteins are expressed (ovals). B. PCR analysis of the multi-cistron cassette genome integration. Transformants were screened by PCR to identify individual clones that correctly integrated the multi-cistronic transformation vector, using the oligonucleotides indicated by the arrows in (A). Three independent clones (cl) are shown. L, ladder; WT, wildtype cc1690; NTC, no template control; P, plasmid. C. Live cell fluorescence microscopy of a clone expressing the multi-cistronic vector. mCerulean-NLS (blue) localizes to the nucleus while BiP-mCherry-HDEL (red) is targeted to the ER. Scale bar, 5 μm.