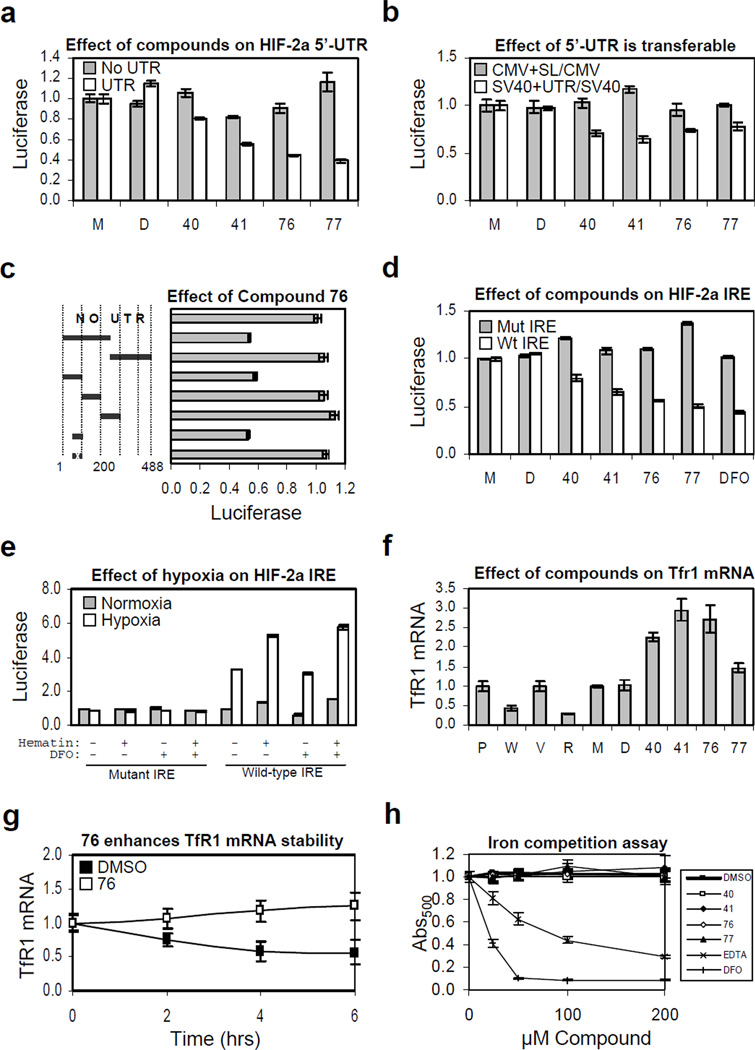
Figure 5. The HIF-2a IRE is necessary and sufficient for mediating the effect of HIF inhibitors and it is de-repressed by hypoxia.
For all panels: P, PRC3; W, WT8; V, pTV; R, pTR, or parental 786-O cells treated with M, medium only; D, DMSO; or compounds, as indicated. Experiments were done in triplicate and error bars represent standard error of the mean (SEM). (a) Effect of compound is dependent upon presence of 5’-UTR. (b) Effect of 5’-UTR is heterologously transferable. Gray bars, ratio of cells expressing CMV-luciferase with a synthetic RNA helicase reporter 5’-UTR stem loop over those expressing luciferase from the CMV promoter alone; white bars, ratio of cells expressing SV40-luciferase with the HIF-2a 5’-UTR over those expressing luciferase from the SV40 promoter alone. Shown are p-values determined using the Student's unpaired homoscedastic t-Test with a two-tailed distribution, comparing the ratios of SV40 driven luciferase with the HIF-2a 5’-UTR over promoter alone versus CMV-driven luciferase with a synthetic stem loop over promoter alone. (c) Effect of compound 76 localizes to IRE element within the 5’-UTR of the HIF-2a message. A series of 5’-UTR deletion mutants were engineered into the HIF promoter driving luciferase. Shown is the normalized ratio of cells treated with 10µM 76 over DMSO only control for each reporter relative to the construct without any 5’-UTR element. The 50 nucleotide sequence with an “x” denotes mutated IRE. (d) All compounds work via the Iron-Responsive Element (IRE). The effect of all compounds was tested on luciferase reporter lines containing wild-type or mutant 5’-UTR IRE element. (e) Hypoxia mediates translational de-repression of the HIF-2a message via the 5’-UTR IRE. Mutant or wild-type HIF-2a IRE luciferase reporter lines were plated in duplicate and treated with medium only, 10µM DFO, 100µM hematin, or both, as indicated and subjected to 24 hours normoxia (gray bars) or hypoxia (white bars). Luciferase measurements were normalized separately for wild-type or mutant IRE to corresponding normoxic reporter line treated with medium only. Raw mutant IRE luciferase activity was roughly 10-fold higher than that of wild-type. (f) Compounds increase expression of Transferrin Receptor 1 mRNA. Quantitative RT-PCR was performed on WT8, PCR3, pTR, and pTV lines as well as compound treated 786-O cells. (g) Compound 76 increases TfR1 mRNA stability. Quantitative RT-PCR showing normalized TfR1 mRNA expression in DMSO versus compound 76 treated 786-O cells following the addition of Actinomycin D. (h) Compounds are not iron chelators. Displacement of iron from ethyl-3,4-dihydroxybenzoate (EDHB) complex was measured as a decrease in absorbance at 500 nm in the presence compounds as well as EDTA and DFO.