Figure 1.
Identification of the Atg1 Kinase Consensus Phosphorylation Site
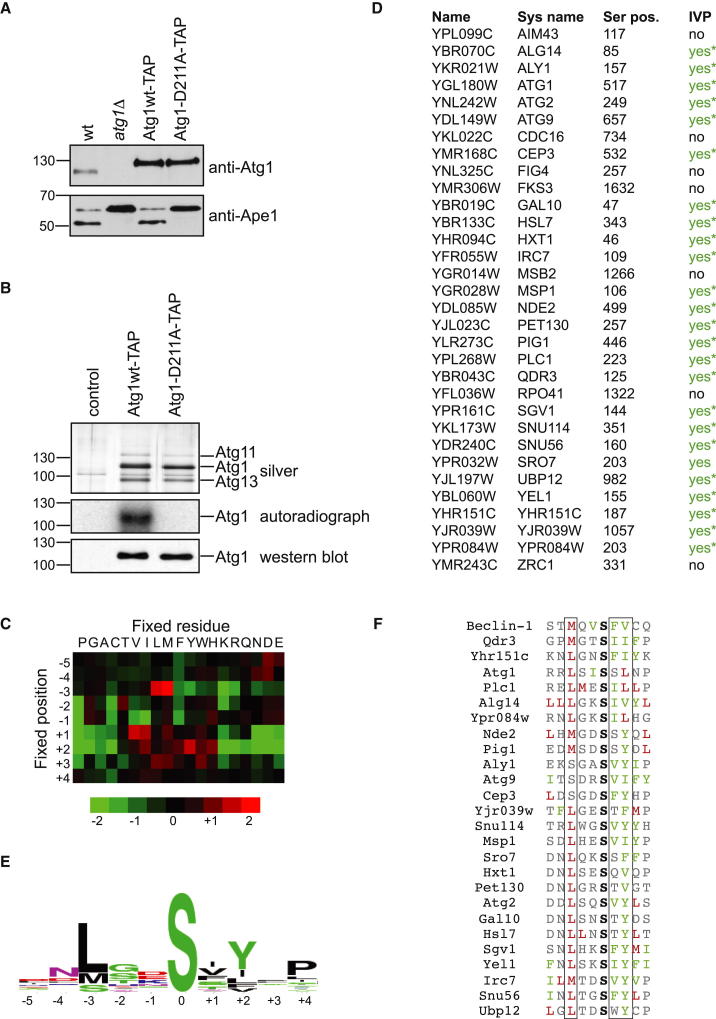
(A) Wild-type (wt), atg1Δ, and genomically integrated TAP-tagged wild-type Atg1 (wt) or Atg1 kinase-dead D211A mutant cells were grown to mid-log phase. Cell extracts were prepared by trichloroacetic acid precipitation and analyzed by anti-Atg1 and anti-Ape1 western blotting. Note that TAP-tagged constructs seem more abundant than untagged Atg1 due to additional antibody binding to protein A in western blotting.
(B) Wild-type (control), genomically integrated TAP-tagged wild-type Atg1 (Atg1wt), or Atg1 kinase-dead D211A mutant cells were grown to mid-log phase and rapamycin treated for 1 hr. Atg1 was immunoprecipitated and eluted by TEV cleavage. The eluate was analyzed by anti-Atg1 western blotting and silver staining, and autophosphorylation activity was assessed in vitro. The identity of bands corresponding to Atg1, Atg11, and Atg13 on the silver gels was confirmed by mass spectrometry.
(C) Peptides (16-mer) containing a central fixed serine phosphoacceptor flanked by degenerate positions and one fixed position as indicated were phosphorylated in vitro using soluble Atg1 complexes, and reactions were spotted on streptavidin-coated membranes followed by autoradiographic quantification. A heat map representing the quantified peptide array is shown.
(D) The quantified consensus motif obtained from the peptide array was searched against the S. cerevisiae protein database. Synthetic peptides spanning the consensus region of the top 30 hits and the consensus region of Atg1 and Atg9 were generated and subjected to in vitro phosphorylation with soluble wild-type Atg1 and the D211A mutant, followed by mass spectrometric identification of their phosphorylation state (IVP). Peptides found to be in vitro phosphorylated are marked with “yes.” Asterisks mark accumulation of precursor ion intensity over time, shown in Figure S5.
(E) A consensus logo was generated by aligning the 25 verified consensus hits using WebLogo (http://weblogo.berkeley.edu) (Crooks et al., 2004).
(F) Alignment of the 25 in vitro verified Atg1 kinase targets with the known ULK1 substrate Beclin-1. M and L are marked in red (position −3 according to the peptide array), and F, V, I, and Y are in green (positions +1 and +2). Note that S can also be found at position −3 and L at position +2. See also Figures S1 and S5.