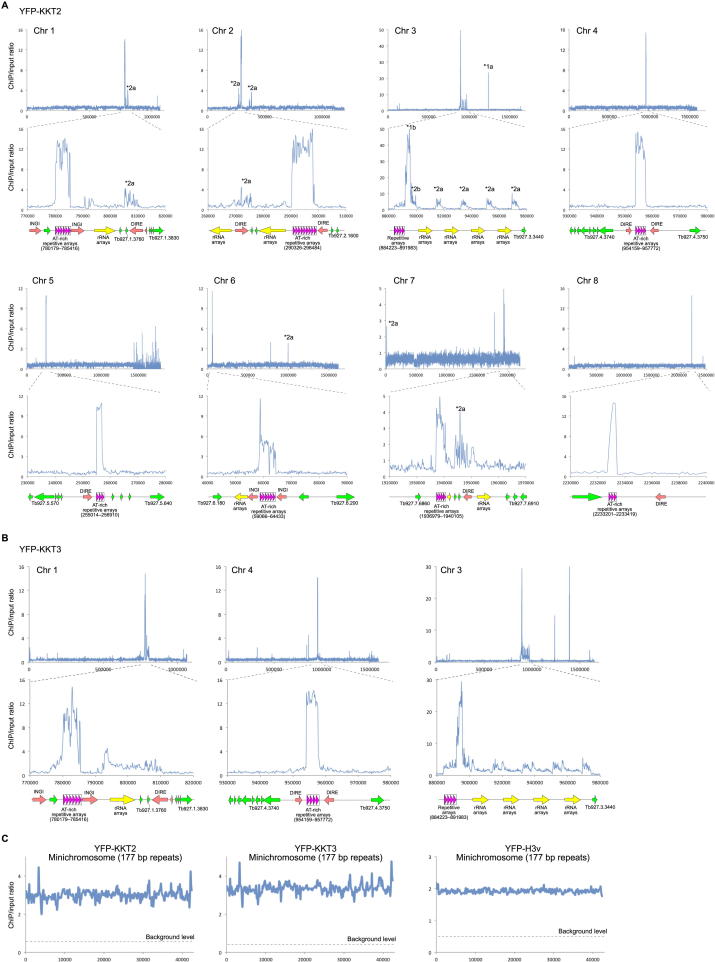
Figure S2.
More ChIP-Seq Data, Related to Figure 2
(A) ChIP-seq data of YFP-KKT2 for chromosomes 1–8 (data for chromosomes 1, 3 and 4 are taken from Figure 2A). Some of the non-centromeric peaks likely represent false-positive signals due to the presence of identical sequences within the centromeres. For example, the ∗1a peak (1,236,842–1,238,050, which is located in between two polycistronic transcription units) is likely false-positive due to the presence of an identical sequence in the centromere of chromosome 3 (∗1b: 892,214–893,422). Similarly, the ∗2a peaks found around some DIRE sequences may be caused by an identical sequence in the centromere of chromosome 3 (∗2b).
(B) ChIP-seq data of YFP-KKT3 for chromosomes 1, 3 and 4.
(C) ChIP-seq data of YFP-KKT2, YFP-KKT3 and YFP-H3v for a model minichromosome that mostly consists of 177 bp repeats. The background level for each protein is indicated by dotted lines.