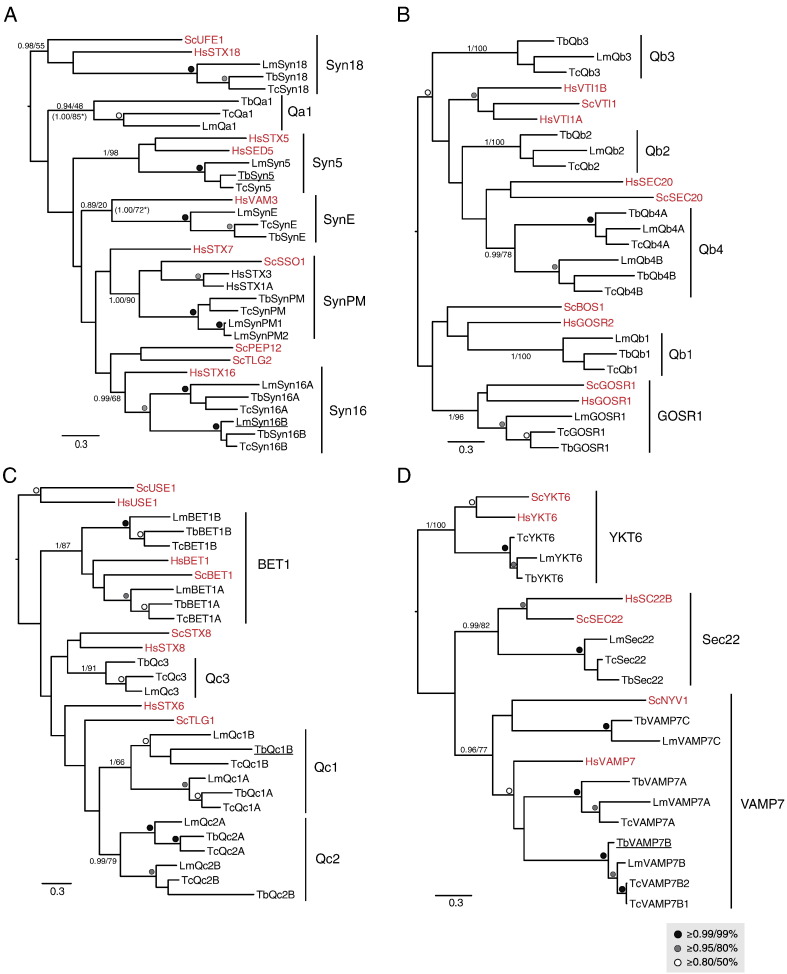
Fig. 1.
Phylogenetic relationships of Trypanosomatid SNAREs. In all panels, the best Bayesian topology is shown, with support values for nodes defining clades of interest given in the order of posterior probabilities (MrBayes) and bootstrap values (PhyML). All values for all other nodes above the threshold of 0.8/50% are iconized as inset. (A) Qa SNARE sub-family analysis. Note the orthology with opisthokont orthologs for Syn18, 5, E, PM, and 16. The Syntaxin16 clade includes two paralogues for each trypanosomatid species. Support values from additional phylogenetic analyses, with long-branching taxa removed, are indicated by asterisks. (B) Qb SNARE sub-family analysis showing orthology with Gos1 and four trypanosomatid Qb clades. (C) Qc SNARE sub-family analysis. Bet1 orthologs plus three additional trypanosomatid clades were reconstructed. (D) R-SNARE analysis. Orthologs for opisthokont sub-families were identified with an expansion in the Vamp7 clades in trypanosomatids. An additional clade of R-SNARE-related trypanosomatid proteins had an unstable position in the phylogeny when the sequences were included (data not shown), therefore, these sequences were removed. Accession numbers for all trypanosomatid sequences are shown in Table S1. Underlined sequences were localized in this study (see Figs. 4–6).