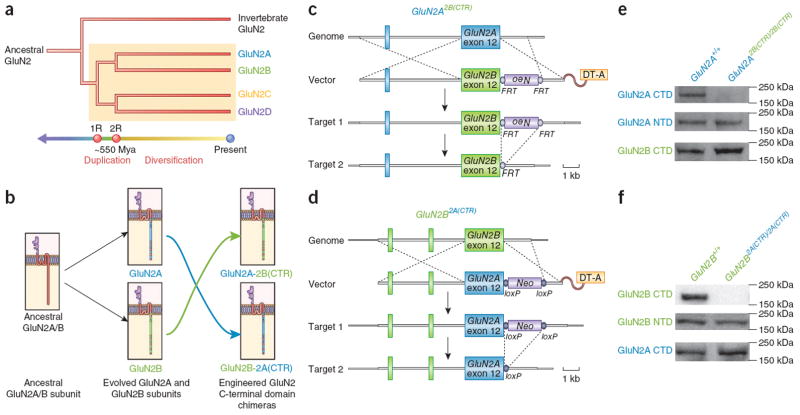
Figure 1.
Design and generation of mouse models a phylogeny illustrating the invertebrate GluN2 and four vertebrate GluN2 paralogs (GluN2A, GluN2B, GluN2C and GluN2D). (a) Two pairs of vertebrate GluN2 genes can be identified, reflecting their evolutionary origins in the two rounds of whole genome duplication (1R, 2R) at the base of the chordate lineage ~550 million years ago (Mya). Yellow box highlights the four vertebrate GluN2 proteins. (b) Schematic depicting the homology (common origin) of the GluN2A and GluN2B subunit. The ancestral subunit to GluN2A and GluN2B was duplicated during the 2R event, creating two GluN2 paralogs that diversified into GluN2A and GluN2B. The cytoplasmic CTDs are the most divergent regions of the GluN2A and GluN2B proteins (Supplementary Fig. 1). CTD regions unique to GluN2A and GluN2B are colored blue and green, respectively. CTD regions that are homologous (common) between GluN2A and GluN2B are colored red (29% of the GluN2A/B CTD amino acid sequences). For brevity, divergent regions of the GluN2A/B extracellular and transmembrane regions are not marked. The engineered GluN2A2B(CTR) and GluN2B2A(CTR) chimeric subunits are depicted beside the respective endogenous murine GluN2 subunits. (c) The GluN2A2B(CTR) allele encodes a chimeric GluN2A subunit consisting of GluN2A extracellular and transmembrane regions and the GluN2B CTD. The terminal GluN2A exon that encodes the CTD was replaced with the paralogous sequence from GluN2B (target 1). The FRT-flanked Neo selection cassette was placed in the GluN2A 3′ UTR and was removed by crossing GluN2A2B(CTR)/+ mice with CAG-FLP recombinase transgenic mice (target 2). (d) The GluN2B2A(CTR) allele encodes a chimeric GluN2B subunit consisting of a GluN2B extracellular and transmembrane regions and the GluN2A CTD. The terminal GluN2B exon that encodes the CTD was replaced with the paralogous sequence from GluN2A (target 1). The loxP-flanked Neo selection cassette was placed in the GluN2B 3′ UTR and was removed by crossing GluN2B2A(CTR)/+ mice with CMV-Cre recombinase transgenic mice (target 2). (e) Western blots of whole forebrain extracted protein, probing for the GluN2A N-terminal domain and CTD, and the GluN2B CTD. No change in apparent protein levels was seen for the GluN2A N-terminal domain in GluN2A2B(CTR)/2B(CTR) mice. No GluN2A CTD signal was detected in GluN2A2B(CTR)/2B(CTR) brain. Probing for the GluN2B CTD revealed an apparent increase in signal in GluN2A2B(CTR)/2B(CTR) mice and a trend toward increased GluN2B CTD signal (normalized to GluN2A N-terminal domain signal) (t4 = 2.9, P > 0.05). (f) Western blots of whole forebrain extracted protein, probing for the GluN2B N-terminal domain and CTD, and the GluN2A CTD. No change in protein levels was seen for the GluN2B N-terminal domain in GluN2B2A(CTR)/2A(CTR) protein extract. No GluN2B CTD signal was detected in GluN2B2A(CTR)/2A(CTR) brain extract. Probing for the GluN2A CTD revealed a significant increase in signal for the GluN2A CTD (normalized to GluN2B N-terminal domain signal) in GluN2B2A(CTR)/2A(CTR) protein extract (t4 = 6.2, P < 0.01).