FIGURE 2.
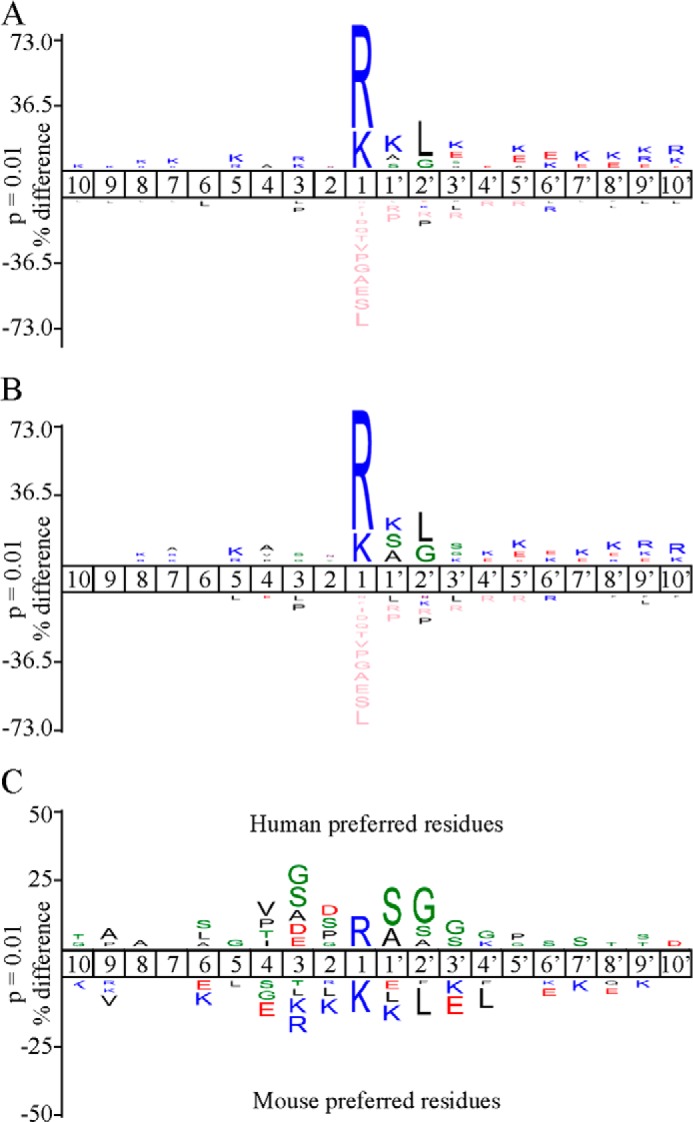
Proteomic analysis of mouse and human granzyme A cleavage specificity. IceLogo (29) visualization of 749 nonredundant mGzmA (A) or 400 nonredundant hGzmA (B) cleavage site motifs identified in the Jurkat proteome. Multiple sequence alignments (29) of peptide substrate motifs from P10 to P10′ were used to identify statistically significant over- or under-representation of residues, relative to the human Swiss-Prot database, with a p value threshold of 0.01. The amino acid heights are indicative of the degree of conservation at the indicated position (pink coloring represent the complete absence of the amino acid indicated). C, differential IceLogo indicating active site specificities based on hGzmA/mGzmA cleavage ratios. Residues cleaved more efficiently by hGzmA are shown above the subsite position, and residues cleaved more efficiently by mGzmA are shown below.
