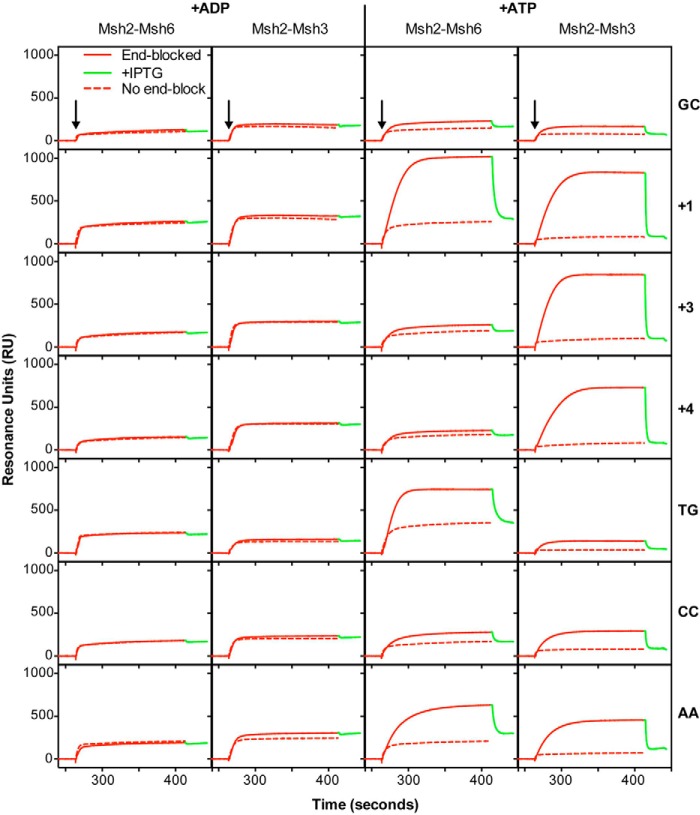
FIGURE 4.
Msh2-Msh3 forms ATP-dependent sliding clamps on substrates containing insertion/deletion mispairs and specific base-base mispairs. The ability of Msh2-Msh3 and Msh2-Msh6 to form sliding clamps on homoduplex DNA or various mispair-containing substrates was monitored using SPR analysis as described under “Experimental Procedures” in the presence of ADP (+ADP) or ATP (+ATP), as indicated. The sensorgrams show the binding to substrates with free ends (dashed red lines) or ends blocked by bound LacI (solid red) followed by release of the end block (green) by the addition of IPTG. The mispair substrate analyzed is indicated at the right of each row of sensorgrams. The x axis indicates the time of association in seconds, the y axis indicates the extent of binding in RUs after reference subtraction, and the black arrows indicate the time at which the injection of protein was initiated.