FIGURE 1.
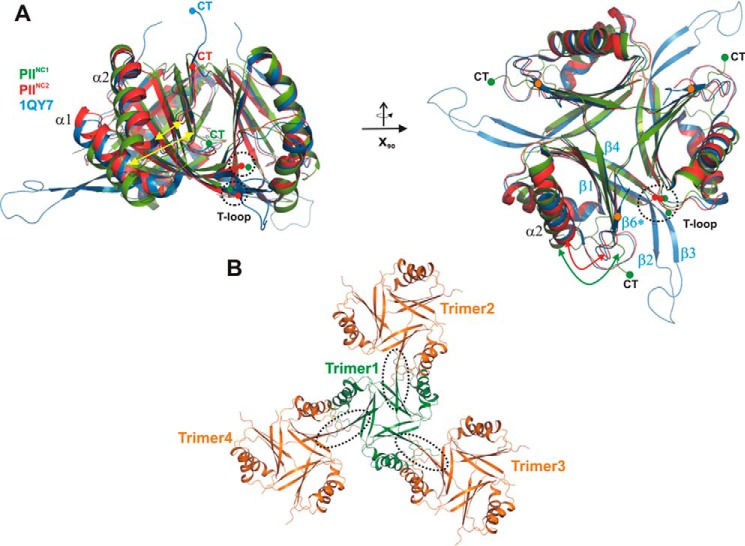
Comparison of PII structures from S. elongatus in the absence of cofactors. All structures are shown as schematics prepared using PyMOL. A, superposition of the two wild-type structures PIINC1 (green) and PIINC2 (red) with the previously published mutant structure (1QY7; blue). The structures are shown in two perspectives (top and side views), which are related to each other through a rotation around the x axis by 90°. The structures show significant deviations (yellow arrows) in the T-loop conformations and the C-terminal extensions (CT), which strongly depend on the crystallographic environment of the protein trimer. B, crystallographic packing of the PIINC1 structure, which shows that the C-terminal β-strand β6 is not formed due to crystallographic contacts (encircled by dotted lines) of trimers via their β5 strands.