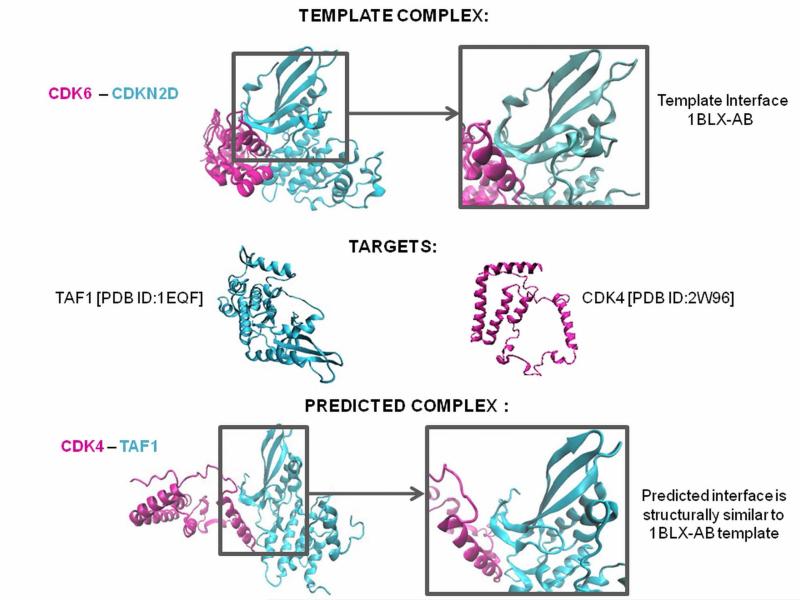
Figure 1. Interface Structure Prediction for Interacting Target Proteins.
Interface information is obtained from the “Protein Interactions by Structural Matching” (PRISM) server. PRISM searches for spatial motif similarity on target proteins’ surfaces using geometric complementarity and considers evolutionary conservation of hot spots based on a non-redundant protein-protein interfaces template dataset derived from the PDB. Its prediction principle is to compare both sides of a template interface with surface regions of any given two monomers, and if they are similar these two proteins are predicted to interact with each other via this interface region. In the above example the CDK6 [PDB:1BLX-A] and CDKN2D [PDB:1BLX-B] complex is derived from PDB and the target proteins CDK4 and TAF1 are found to be interacting via an interface structurally similar to 1BLX-AB interface. CDK 4 and TAF1 are predicted to be interacting via 1BLX-AB interface.