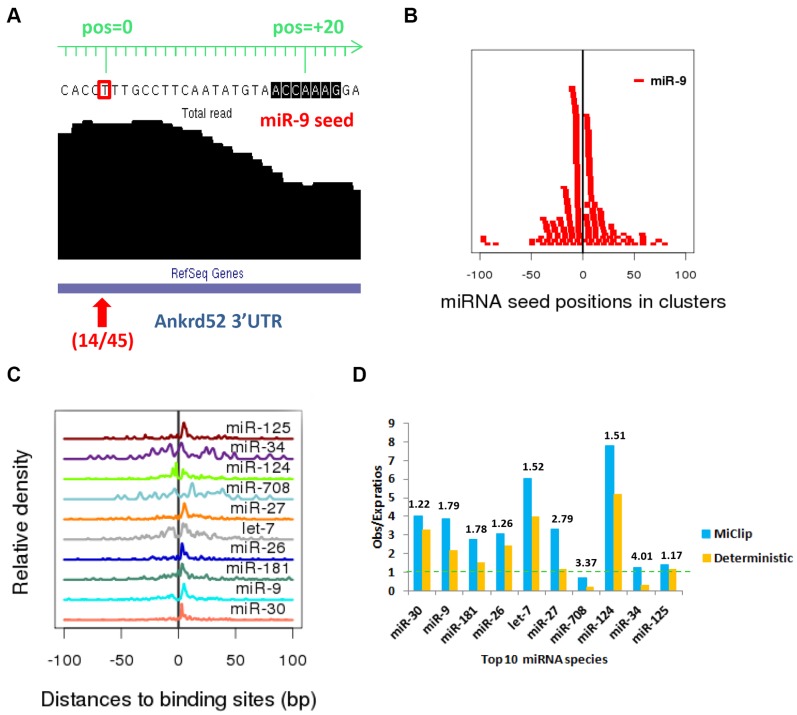
Figure 3. miRNA seed matches in AGO mRNA CLIP clusters.
(a) An exemplary cluster with one reliable binding site and a seed match for miR-9. The red rectangle is the binding site, which has 14 mutant tags out of 45 total tags. The miR-9 seed motif is ACCAAAG, which locates 20 bp downstream of the binding site. (b) The positions of seed matches of miR-9 (positions 2–8) within the 5,795 significant clusters. 4 out of 199 seed motifs are more than 100 bp upstream or downstream away from the binding sites, which are ignored. X-axis is the relative position with respect to the binding site within each cluster. (c) The pileup of seed matches of the top 10 miRNAs (positions 2–8) within the 5,795 significant clusters drawn as density curves. 49 out of 1557 seed motifs (all top 10 miRNAs) are more than 100 bp upstream or downstream away from the binding sites, which are ignored. (d) The signal/noise ratio of miRNA seed motif enrichment. The blue bars show enrichment ratios for MiClip and the orange bars show enrichment ratios for the ad hoc method. The numbers on top of these bars show the signal/noise ratio for clusters found by MiClip divided by the signal/noise ratio for clusters found by the ad hoc method.