Figure 3. Luciferase assays for the Twist2 locus using constructs where the ZBED6 binding site was altered.
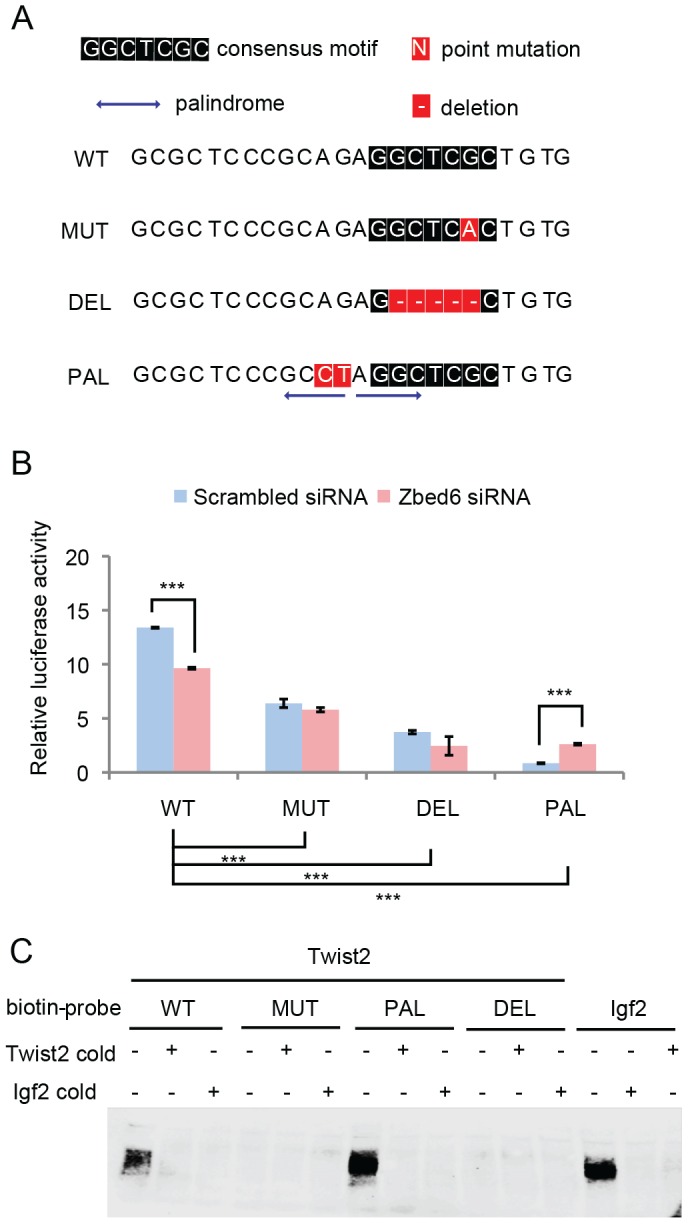
(A) The 24-bp conserved ZBED6 binding site at the Twist2 gene (WT) was mutated to generate MUT containing a single G-to-A mutation at the ZBED6 consensus motif, to DEL by deleting the consensus motif, and to PAL by altering two nucleotides that created the same palindrome sequence as present at the Igf2 locus (PAL). (B) The relative luciferase activity (the ratio of firefly to Renilla) of WT, MUT, DEL, PAL constructs in Zbed6-silenced (blue) or control C2C12 cells (pink). Standard errors of the mean are displayed by the error bars. ***, P<0.001. (C) EMSA of WT, MUT, DEL, PAL and Igf2 biotin probes incubated with C2C12 nuclear extracts showing binding of endogenous ZBED6 to the WT, PAL and Igf2 biotin probes but not to the MUT and DEL biotin-probes. The binding complex was competed using an 100-fold excess of the corresponding Twist2 cold probe or Igf2 cold probe.
