Abstract
Quorum sensing, a form of cell–cell communication among bacteria, allows bacteria to synchronize their behaviors at the population level in order to control behaviors such as luminescence, biofilm formation, signal turnover, pigment production, antibiotics production, swarming, and virulence. A better understanding of quorum-sensing systems will provide us with greater insight into the complex interaction mechanisms used widely in the Bacteria and even the Archaea domain in the environment. Metagenomics, the use of culture-independent sequencing to study the genomic material of microorganisms, has the potential to provide direct information about the quorum-sensing systems in uncultured bacteria. This article provides an overview of the current knowledge of quorum sensing focused on phylogenetic diversity, and presents examples of studies that have used metagenomic techniques. Future technologies potentially related to quorum-sensing systems are also discussed.
Keywords: N-acyl-L-homoserine lactone, metagenomics, quorum quenching, quorum sensing, quorum-sensing inhibitor, quorum-sensing signal
Microbial Communication: Quorum Sensing
Bacteria interact with one another using chemical molecules as sensing signals. Detection of the molecules allows bacteria to distinguish between low and high cell-population densities and to control gene expression in response to changes in cell number1 and the local environment.2 This process, referred to as quorum sensing (QS), allows a population of bacteria to coordinately control gene expression. Several types of QS signals have been found, including N-acyl-L-homoserine lactone (AHL) in the Proteobacteria.1,3,4 In these bacteria, AHL-dependent QS systems have been shown to regulate many bacterial behaviors, such as virulence5,6 and biofilm formation,7,8 mainly in response to cell densities. AHL-dependent QS systems are now recognized to play important roles in the regulation of bacterial behavior. The first QS system to be discovered was the Lux system of Vibrio fischeri, which regulates light production in the light organs of certain deep-sea fish and squid using AHL-signaling molecules. AHL is composed of a homoserine lactone (HSL) ring with an acyl chain (Fig. 1A). The acyl-chain length typically varies from C4 to C12 and may be modified by a 3-oxo substituent or a 3-hydroxy substituent or a degree of unsaturation. The AHL-based QS systems usually contain a luxI gene homolog, which is responsible for the synthesis of AHLs, and a luxR gene homolog, which acts as an AHL-responsive transcriptional regulator.9 When the number of cells reaches a given threshold or bacterial “quorum”, the AHL concentration becomes sufficiently high for the signaling molecules to be recognized by an AHL-responsive transcriptional regulator, LuxR, which activates or represses target genes.
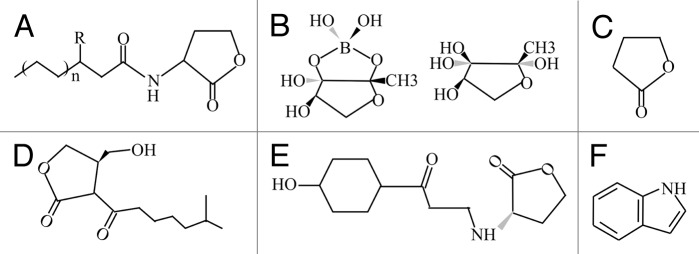
Figure 1. Chemical structures of representative QS signals. (A) N-Acyl-L-homoserine lactone (AHL), R represents either the 3-oxo substituent or absence of substitution. (B) Autoinducer-2 (AI-2), (C) γ-Butyrolactones, (D) A-factor, (E) p-Coumaryl homoserine lactone (pC-HSL), (F) Indole.
LuxI catalyzes the formation of the AHL from the substrates S-adenosyl-l-methionine (SAM) and acyl–acyl carrier protein (acyl-ACP).10 The LuxI enzymes are composed of a single domain of approximately 205 amino acid residues in length. The LuxI sequence is conserved among members of the AHL synthase family, which share more than 20% identity/40% similarity within this sequence.11 The conserved region and conserved residues are apparent in an alignment of the AHL synthases (Fig. 2). The N-terminal region of the LuxI sequence is the most conserved part of the enzyme. This region includes invariant residues that are crucial for the enzymatic activity, and that play a role in the binding of the conserved substrate, SAM, and catalysis.12 The C-terminal region of the enzyme is less conserved overall, since this region is involved in recognition of the variable part of the acyl-ACP substrate, which has an acyl chain of varying length.
Figure 2. Multiple alignments of amino acid sequence of LuxI family members. Amino acid sequences were aligned using ClustalW. Amino acid residues conserved among all members are highlighted in black. Asterisks indicate the 18 amino acid residues comprising the binding tunnel in LasI. Identical amino acid residues with LasI are identified by a gray background. Brackets beside the sequences indicate three groups classified by carbon chain length of the synthesized AHL. The threonine residue, which is essential for determining the specificity of the enzyme for 3-oxo-acyl-ACP, is shown by a box. The amino acid sequences used in the alignment are LasI (GenBank No. P33883), PpuI (AAM75411), QS6–1 (ACH69662), QS10–1 (ACH69667), QS10–2 (ACH69672), MupI (AAK28505), YenI (CAA53693), SinI (CAC46418), TraI (AAB95104), CepI (AAD12727), YpsI (AAD40486), LuxI (CAA68562), CarI (P33880), EsaI (AAA82096), QS1 (AAT90822), PhzI (AAC41535), RhlI (AAC44037), AsaI (AAB70017), and AhyI (ABD59318).
LuxR is the most studied member of the newly emerging family of transcriptional regulators involved in AHL-mediated quorum sensing. A 20-bp inverted repeat sequence, which was found close to the −35 box of sigma 70 promoter is involved in the activation of LuxR. This inverted repeat is typically considered to be a lux box-like element where a LuxR regulator binds to the LuxI/LuxR-type QS systems.13-15 The LuxR homolog LasR was discovered in P. aeruginosa.16 LasR protein act in concert with AHL to coordinate the expression of target genes, including genes encoding virulence factors. TraR from Agrobacterium tumefaciens binds to AHL and regulates conjugal plasmid transfer,17 and ExpR from Pectobacterium carotovora regulates exoenzyme production and antibiotic synthesis in this bacterium. All LuxR homologs have a low similarity with LuxR (25% identity).
LuxI and LuxR homologs have been identified in diverse species throughout the gram-negative Proteobacteria, including phototrophic purple non-sulfur bacteria, marine vibrios, rhizosphere bacteria, enteric commensals, and opportunistic pathogens of plants and animals. To our knowledge, AHL-producing bacteria have been identified in over 37 genera within the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria,6,18-20 and Cyanobacteria.21 The LuxI/LuxR-type QS systems have been experimentally characterized in more than 70 different species in the phylum Proteobacteria.22,23 Genome sequencing of a number of cultured Deltaproteobacteria24,25 and a yet-to-be cultured bacterium belonging to the phylum Nitrospirae26 indicates that they may harbor putative LuxI/LuxR-type QS systems. It has been speculated that half the bacterial phyla (26 candidate phyla) do not have cultivated representatives, although at least 52 bacterial phyla have been identified from 16S rRNA gene sequences in environmental samples.27 These results suggest that not only Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria, but also a diverse array of other bacteria, including as-yet-uncultivated bacterial phyla, likely possess LuxI/LuxR-type QS systems. More recently, a bacterial LuxI/LuxR-like QS system was found in a methanogenic archaeon, Methanosaeta harundinacea 6Ac, and shown to be involved in regulating cell assembly and carbon metabolic flux.28
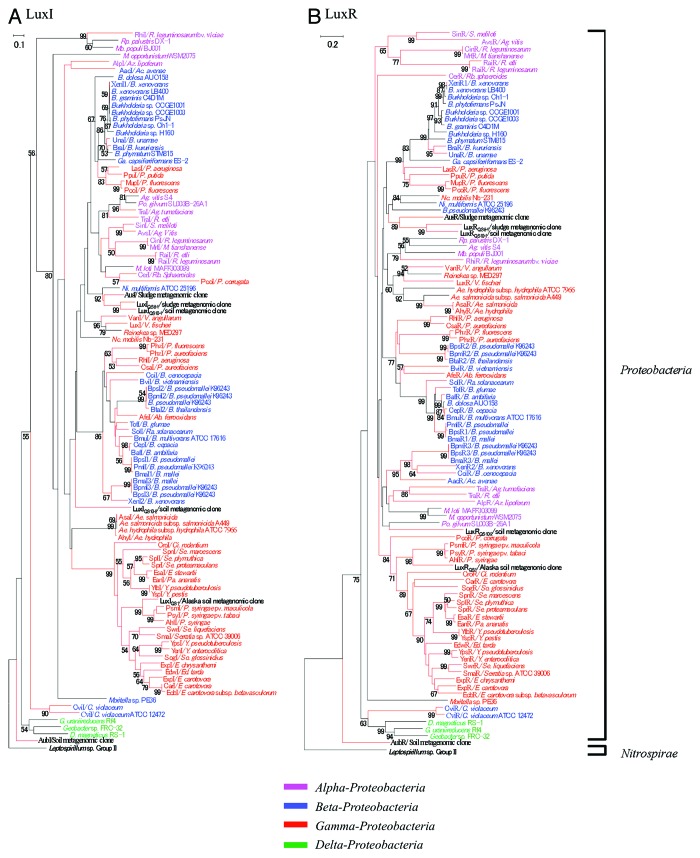
Based on the neighbor-joining trees generated from aligned LuxI and LuxR sequences, the majority of proteins in each family are clustered within the same groups defined by 16S rRNA sequences of bacteria29 (Fig. 3). Although individual exceptions to this rule were observed, the overall congruity between the quorum sensing and the rRNA trees is consistent with the notion that the quorum-sensing proteins within the Proteobacteria are of ancient origin.
Figure 3. Phylogenetic trees of (A) LuxI and (B) LuxR protein sequence homologs obtained from Proteobacteria and Nitrospirae. The members isolated from metagenomic clones are indicated in bold type. The characterized proteins AubI, AubR, AusI, and AusR are highlighted by red closed boxes. Species belonging to the α-, β-, γ-, and δ-Proteobacteria are grouped within brackets. A Leptospirillum sequence is used as an outgroup. The bold brackets represent two groups of phyla. The branch length and bootstrap values were obtained from 1000 replicates using a neighbor-joining algorithm. Nodes lacking a number are those that could not be supported by more than 50% of the bootstrap value. The scale bars represent 0.1 and 0.2 substitutions per amino acid site, respectively. The red branches are sequences with experimental evidence. Abbreviations for the bacterial genus names are as follows: Ab, Acidithiobacillus; Ac, Acidovorax; Ae, Aeromonas; Ag, Agrobacterium; Az, Azospirillum; B, Burkholderia; C, Chromobacterium; Ci, Citrobacter; D, Desulfovibrio; E, Erwinia; Ed, Edward; G, Geobacter; Ga, Gallionella; M, Mesorhizobium; Mb, Methylobacterium; Ni, Nitrosospira; Nc, Nitrococcus; P, Pseudomonas; Pa, Pantoea; Po, Polymorphum; R, Rhizobium; Ra, Ralstonia; Rb, Rhodobacter; Rp, Rhodopseudomonas; S, Sinorhizobium; Se, Serratia; So, Sodalis; V, Vibrio; Y, Yersinia.
Metagenomics: Access to Uncultured Bacteria
A better understanding of QS systems will provide us with greater insight into the complex interaction mechanisms among the Bacteria and even the Archaea in the environment. Previous research has been limited by the lack of information on uncultivated members of the community. More than 99% of the microbes that exist in the environment cannot be cultivated easily,27,30 and it has been reported that approximately 107 cells are present in 1 g of soil.31 Thus, most of the microbes in the environment have not been described or accessed for biotechnology or basic research. These uncultured microorganisms represent potentially important sources for the discovery of novel genes, including QS systems that catalyze the production of novel QS signals. “Metagenomics”, which is the culture-independent genomic analysis of the microbial community, attempts to overcome these difficulties.32,33 This technology has the potential for providing insight into the functional dimensions of environmental genomic data sets and will help to achieve a major goal of environmental microbiology—that is, clarifying the complexities of microbial community function and the interaction among these microbes.
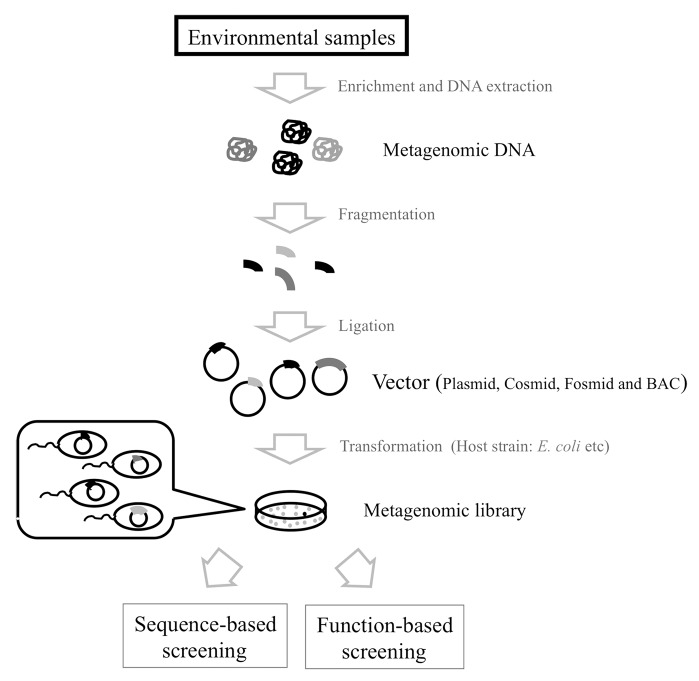
Metagenomic analysis is usually initiated by the isolation of DNA from environmental samples, and then this DNA is used to construct a metagenomic library by cloning DNA fragments into an appropriate vector (plasmid, cosmid, fosmid, etc.), or used directly for high-throughput sequencing such as 454 pyrosequencing (Fig. 4). To increase the efficiency of gene isolation in metagenomic samples, many laboratories isolate metagenomic DNA from a microbial community after enrichment in the laboratory. To construct a metagenomic library from the purified DNA, Escherichia coli has been used as a host strain. Construction of the metagenomic library is followed by screening for novel genes. Two major strategies have been used to identify genes encoding novel enzymes and the production of chemical compounds. One of these methods, the function-based screening method, is generally selected based on the enzymatic activity of novel gene products in the metagenomic library.34-36 In this method, environmental DNAs are cloned into expression vectors and propagated in appropriate hosts, followed by evaluating the activity expressed by the recombinants.37-40 The evaluation of activity is often performed using a sensor strain or an indicator substrate that shows a color change as a result of biocatalytic conversion on agar plates. Because it is not dependent on previously sequenced genes, this method has the potential to identify novel classes of functional enzymes.
Figure 4. Construction and analysis of metagenomic libraries from environmental samples. The metagenomics involved constructing a DNA library and analyzing the functions and sequences in the library.
Screening for LuxI/LuxR-Type QS Quorum-Sensing System from a Metagenomic Library
By using a function-based metagenomic screening approach, new AHL synthase genes have been identified from uncultured organisms.41,42 A high–throughput “intracellular” screening system has been designed and used to screen for quorum-sensing signals from metagenomic libraries. Williamson et al. have designed an intracellular screening system, designated METREX, in which metagenomic DNA is cloned in a bacterial host containing a biosensor for compounds that induce bacterial quorum sensing.41,43 If the metagenomic clone produces a quorum-sensing inducer, the bacterial cell produces green fluorescence protein (GFP) and can be identified by fluorescence microscopy or captured by fluorescence-activated cell sorting. In a study using the METREX method, screening of metagenomic libraries constructed from Alaskan soil led to the discovery of a novel LuxI/LuxR-type QS system with low similarity to the known homologs found in Gammaproteobacteria.41 In another study, metagenomic analysis of the gypsy moth gut microbiota using the METREX method identified a gene that encodes a monooxygenase homolog that mediates production of signal mimics that induce QS.44
Hao et al. used an A. tumefaciens biosensor strain HC103 (pJZ381) to screen metagenomic libraries for new QS inducers. When metagenomic clones are introduced into the biosensor strain, if quorum-sensing inducer synthases are present, the active signals produced bind to TraR and activate transcription of the traC–lacZ fusion gene, resulting in blue colonies in the presence of 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-Gal). The analysis of metagenomic libraries constructed from activated sludge and soil resulted in the isolation of three new LuxI/LuxR-type QS systems producing novel AHLs most closely related to those previously found in Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria.42
Furthermore, Nasuno et al. used a metagenomic approach to find novel LuxI/LuxR-type QS systems in uncultured bacteria belonging to classes other than the previous studied Proteobacteria, such as the phylum Nitrospirae. In this study, the metagenomic libraries were constructed from the activated sludge of a coke plant wastewater treatment system containing various organic pollutants39,45 and forest soil as a source of phylogenetically diverse microbes. An “intercellular” screening method was used, and GFP fluorescence of the biosensor was detected using a spectrofluorometer (Fig. 5). DNA sequence analysis revealed two pairs of new LuxI family N-acyl-L-homoserine lactone (AHL) synthases and LuxR family transcriptional regulators (designated AubI/AubR and AusI/AusR, respectively). Phylogenetic analysis based on amino acid sequences suggested that AusI/AusR was from an uncultured member of the Betaproteobacteria, whereas AubI/AubR was very deeply branched from previously described LuxI/LuxR homologs in Proteobacteria, indicating that it was not acquired recently from proteobacteria but rather has a more ancient ancestry (Fig. 3). These results demonstrated that metagenomic approaches are useful for the discovery of novel QS systems from uncultured bacteria.
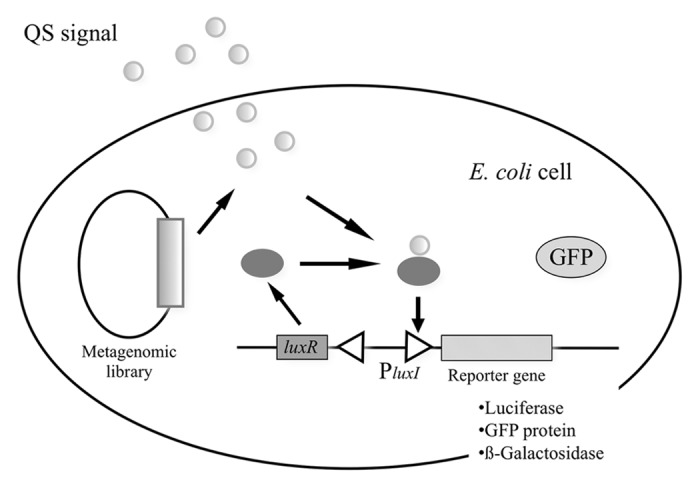
Figure 5. Biosensor system designed for detection of AHL and AHL mimics. In this intracellular screen, the biosensor that detects active clones is inside the same cell as the metagenomic DNA. In the case of the intercellular screen, the biosensor that detects active clones is in a different cell than the metagenomic DNA. The biosensor detects small diffusible signal molecules that induce quorum sensing. When the signal molecules reach a sufficient concentration, they bind the LuxR transcriptional activator that activates the luxR promoter and induces the expression of target genes, in this case GFP. Luciferase and β-galactosidase genes have been also used as reporter genes.
A Novel Quorum-Sensing Signal Isolated by Metagenomic Screening
Several QS signals, including AHL, have been discovered in cultured microorganisms (Fig. 1). Autoiducer-2 (AI-2) is a furan-like quorum-sensing signal that was initially identified from Vibrio harveyi, and its synthesis is catalyzed by a luxS gene that has been found in over 70 bacterial species.3,46 γ-Butyrolactone is used as a signal compound to control morphological differentiation and secondary metabolite production in Streptomycetes. Although γ-butyrolactones are structural analogs of AHLs, there is no known overlap in synthesis or cross-talk between the two signals. A-factor, which is produced by Streptomyces griseus, has been well-studied and antagonizes a DNA-binding repressor protein, thereby promoting the production of the antibiotic streptomycin.47 In addition to these compounds, a number of other QS signals are still being identified, such as p-coumaryl homoserine lactone (pC-HSL), which is classified as a structurally novel member of the AHLs.48
Indole is formed from tryptophan by tryptophanase and can be transformed to various oxidative compounds by bacterial enzymes. Recently, several studies have revealed that indole plays diverse biological roles as a signal molecule in bacteria (Fig. 1). Wang et al. have demonstrated that indole activates the transcription of genes encoding tryptophanase and enzymes required for amino acid degradation in E. coli.49 A family of indoles regulates virulence and shiga toxin production in pathogenic E. coli. Virulence of P. aeruginosa is also controlled by indole.50-52 The effects of indole on biofilm formation in Vibrio cholerae,53 non-pathogenic E. coli K-12, and pathogenic E. coli O157:H754,55 have also been investigated. Previous studies suggest that there are numerous unknown compounds, including indole derivatives, that can interfere with or activate AHL or other signal-mediated QS systems. These structurally similar compounds seem to affect each other as signaling molecules or just as antagonists in the natural environments. The actual roles of these compounds in physiology have not been rigorously investigated, and the discovery of novel bioactive compounds and their functions in the natural environment will lead to real understanding of the chemical interactions among all forms of life.
In previous studies, the discovery of QS signals was accomplished by chemical purification and structural identification of signal compounds from the cultivated bacteria. Therefore, these studies were limited by a lack of information on uncultivated members of the community. Recently, a metagenomic screening method was applied to the search for novel signal compounds. Chemical analysis revealed that the signal mimics were identified as uncharacterized indole derivatives from gypsy moth gut metagenomic library. Guan and coworkers proposed that these signals involving the production of blue-colored compounds represent a new structural class of QS signals, since the compounds are chemically distinct from previously described QS inducers. However, the structure of these indole-based and indoxyl derivatives could not be determined due to their instability during purification. Future studies will be needed to clarify the chemical structure of these new signals and their biological role in the microbial community.
Metagenomic Screening for Quorum-Sensing Inhibitors
AHL-dependent QS regulates many bacterial behaviors, such as virulence56-59 and biofilm formation.60,61 Therefore, AHL-dependent QS is increasingly seen as a useful target for controlling bacterial behaviors. The most obvious way to quench QS-mediated gene expression is to inactivate the signaling molecule itself to prevent it from accumulating. Indeed, a wide variety of AHL-inactivating bacteria belonging to Acinetobacter (Gammaproteobacteria), Agrobacterium (Gammaproteobacteria), Comamonas (Betaproteobacteria), Ochrobactrum (Alphaproteobacteria), Pseudomonas (Gammaproteobacteria), Ralstonia (Betaproteobacteria), Shewanella (Gammaproteobacteria), Variovorax (Betaproteobacteria), Tenacibaculum (Bacteroidetes), and Nostoc (Cyanobacteria) have been isolated and characterized.62-73 Furthermore, AHL-inactivating Gram-positive bacteria within the Arthrobacter, Bacillus, Microbacterium, Nocardioides, Rhodococcus, and Streptomyces genera are also found to have the ability to inactivate AHLs, suggesting that phylogenetically diverse bacteria can quench the AHL-based QS.66,68,74-78
AHL-inactivating enzymes have been found in AHL-inactivating bacteria, and these enzymes can be divided into three representative families: AHL-lactonases, AHL-acylases, and AHL-oxidoreductases (Fig. 6). The prototype AHL lactonase was cloned from Bacillus sp. 240B1 and shown to hydrolyze the ester bond of the lactone ring to give acylhomoserine.74 Furthermore, Uroz et al. reported the isolation of the gene (qsdA) encoding an AHL-inactivating enzyme from Rhodococcus erythropolis strain W2.79 The qsdA gene encodes a phosphotriesterase (PTE)-like broad-spectrum AHL lactonase, which is found only in the Rhodococcus genus and solely in strains capable of degrading AHLs. On the other hand, a strain of Variovorax paradouxus was found to degrade AHLs by an acylase in which the amide bond connecting the HSL ring to the acyl chain is cleaved, releasing homoserine lactone and a fatty acid, which is further metabolized. AHL-oxidoreductase activity was first observed from R. erythropolis, which reduces AHLs with 3-oxo substituents to their corresponding 3-hydroxy derivatives.80 Although the AHL-oxidoreductase gene has not been cloned yet, an oxidoreductase protein from R. erythropolis was purified and characterized.81,82 Database searches for homologs of the characterized AHL lactonases and acylases in complete bacterial genomes have shown that relatives of these enzymes exist in a wide range of species, indicating that bacteria that possess these activities may be widespread in the environment.83
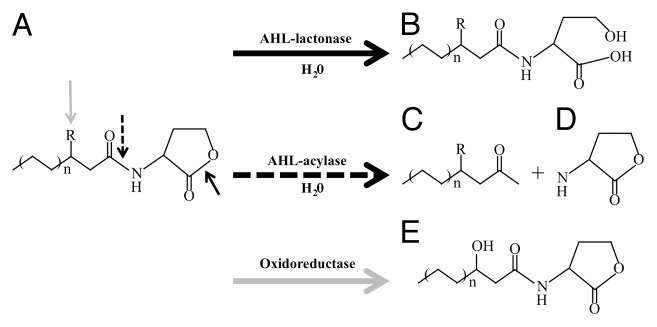
Figure 6. Schematic representation of the enzymatic reactions of various quorum-sensing signal degradation and modification enzymes. AHL signal inactivation, where R represents either a 3-oxo substituent or absence of substitution, is shown. (A) AHL family, (B) acyl-homoserine, (C) fatty acid, (D) homoserine lactone, (E) 3-hydroxy AHL.
With the widespread appearance of antibiotic-resistant bacteria, there is an increasing demand for novel strategies to control infectious diseases. Although the role of AHL-inactivating enzymes in bacteria has not been fully resolved, the AHL-inactivating enzymes may have utility as therapeutic agents to inhibit microbial virulence and pathogenicity. To exploit the novel types of QQ (quorum quenching) enzyme from the uncultured bacteria, a metagenomic approach has been used to isolate the genes encoding AHL-inactivating enzymes. A metagenomic library generated from bacteria inhabiting pasture soil in France was screened for the presence of fosmid clones conferring QQ ability upon their E. coli host.84 One clone demonstrated QQ activity, and a gene, qlcA, encoding an AHL-lactonase activity was identified. The QlcA protein belongs to the family of zinc-dependent metallohydrolases and appears to be distantly related to other AHL-lactonases discovered in Agrobacterium, Bacillus, Photorhabdus, and Rhizobium species. In addition, a metagenomic approach allowed the discovery of a novel bacterial QQ enzyme (QsdB) in a rhizosphere that was treated with γ-caprolactone.85 QsdB is the first amidase signature (AS) family member exhibiting QQ-activity. These results indicate that a metagenomic approach is a useful tool for identifying a variety of QQ enzymes from environmental samples.
In addition to the metagenomic screens to access the unculturable bacteria, screens of QS inhibitors (QSIs) have been attempted from cultivated bacteria in order to develop new drugs for biofilm-based chronic infections.86-89 Lee et al. have reported that biofilm formation was inhibited by 7-hydroxyindole and 5-hydroxyindole in E. coli.90 It has also been reported that 4-hydroxyindole, 5-hydroxyindole, 6-hydroxyindole, and isatin inhibited not only membrane vesicle production but also Pseudomonas quinolone signal (PQS) synthesis.91 These derivatives were suggested to either act as bacterial signals or antagonists of indole, or to inhibit the binding of indole to the signal receptor. Recently, it has been revealed that one of the halogenated furanone derivatives, (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone from a marine macroalgae, inhibits the QS systems in E. coli K-12.92 Although previously reported QSIs have been isolated from the cultivated bacteria, the microbial community may be a target of research to discover novel QSI compounds.
Concluding Remarks and Future Prospects
Metagenomic technology is currently applied to a broad range of tasks, such as determining the potential applications of QS systems, QS signals, and QS inhibitors. Metagenomic technology is able to study microorganisms in the environment that are, as yet, unculturable, and which represent more than 99% of the organisms in some environments. It is expected that the number of novel QS systems identified using metagenomic technology will exceed the number identified through sequencing of QS systems from isolated individual microbes. Metagenomics may provide insight into the functional dimensions of QS systems in microbial communities and will help to achieve a major goal of microbiology: the linking of individual microbial species to function.
However, metagenomics is still a developing technology with limitations to be overcome. A metagenomic approach has a significant limitation in terms of the frequency with which QS system genes are identified (the hit rate). Statistically, for a small-insert (<10 kb) library, between 105 and 106 clones need to be screened for a single hit.93 These results suggest that the discovery of QS system genes in a complex metagenome would be technically challenging. The hit rate depends on a combination of the following factors: the choice of a vector, the assay method, and the efficiency of heterologous gene expression in a surrogate host. Of these factors, the efficiency of gene expression is considered to be one of the most critical. Lämmle et al. have developed a plasmid vector with a dual-orientation promoter to double the chances of transcribing the cloned insert fragments, and this system succeeded in increasing the hit rate about 10-fold.94 However, many genes, perhaps most, will not be expressed in any particular host bacterium selected for cloning, such as E. coli. To increase the capability of identifying the target genes that are difficult to express in E. coli, Streptomyces lividans, and Pseudomonas putida were developed as initial alternative hosts. S. lividans was shown to be a particularly useful host for functional screening of soil metagenomic libraries for novel polyketide synthase genes.95 In addition, Craig et al. showed that other bacterial species, including Burkholderia graminis and Agrobacterium tumefaciens, were also very useful hosts for the expression of environmental genes.40 In addition, the choice of a vector depends on whether the target of study is individual genes or entire operons and gene clusters encoding QS systems. To clone the entire gene clusters containing the QS system genes, the construction of large-insert libraries is required. Among several available vectors, plasmids (suitable for cloning smaller than 10 kb DNA fragments) have high copy numbers and strong vector-borne promoters, but cannot improve the hit rate compared with other low copy vectors, such as cosmids (25–35 kb), fosmids (25–40 kb), and BACs (100–200 kb).33 An additional technique that includes an enrichment step for microorganisms harboring the desired traits also needs to be employed before library construction to increase the hit rate.96-98 Although the enrichment step results in a reduction of microbial diversity, the combination of enrichment and metagenomic is useful for increasing the number of positive clones in a screen and isolating novel bioactive compounds from complex habitats.37,98
The assay methods will also improve to increase the hit rate in the activity-based screening. The assays for detecting QS systems are usually performed by using sensor strains. These strains have the ability to produce GFP protein, luciferase, and β-galactosidase when the QS signal is present. The agar plate assay requires no special devices and can be performed at high throughput, since positive clones can be distinguished by visual inspection based on their color. However, the strength of the output signals is often significantly weak, which could be one of the reasons for the low hit rate. To improve the detection sensitivity of the assay, some alternative approaches have been applied. Using liquid cell lysates of metagenomic clones in a microtiter-plate format is one strategy for increasing the screening sensitivity.99 The biosensor responded with a signal compound producing from a metagenome clone could be sensitively detected using a spectrofluorometer based on the GFP fluorescence. Crude cell lysates were prepared by either chemical99,100 or physical101 procedures. The cells are lysed by addition of a protein extraction reagent in the chemical procedure, while the cells are broken in a vibrating crusher using glass beads in the physical procedure. The detection sensitivity of enzymatic activity could be dramatically improved using bacterial cell lysates instead of intact cells. To identify unknown QS signals from the metagenomic library, the development of a sensor strain is an important goal.
Furthermore, next-generation sequencing (NGS) is becoming one of the standard high throughput data acquisition techniques that has been applied to both genomics and metagenomics. The large amounts of data derived from next-generation sequencing will be appropriate resources for building an efficient data mining strategy. A website and database named Quorumpeps® have been developed for analyzing QS system information.102 Quorumpeps® is a resource for information on quorum-sensing signaling peptides, including information about the structure, activity, physicochemical properties and related literature. All resources are freely accessible at http://quorumpeps.ugent.be/.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Glossary
Abbreviations:
- QS
quorum sensing
- AHL
N-acyl-L-homoserine lactone
quorum quenching
- QSI
quorum-sensing signal
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/27850
References
- 1.Williams P, Winzer K, Chan WC, Cámara M. Look who’s talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond B Biol Sci. 2007;362:1119–34. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decho AW, Norman RS, Visscher PT. Quorum sensing in natural environments: emerging views from microbial mats. Trends Microbiol. 2010;18:73–80. doi: 10.1016/j.tim.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–86. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 4.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 5.Xu L, Li H, Vuong C, Vadyvaloo V, Wang J, Yao Y, Otto M, Gao Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2006;74:488–96. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton EO, Read HW, Pellitteri MC, Hickey WJ. Identification of acyl-homoserine lactone signal molecules produced by Nitrosomonas europaea strain Schmidt. Appl Environ Microbiol. 2005;71:4906–9. doi: 10.1128/AEM.71.8.4906-4909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labbate M, Queck SY, Koh KS, Rice SA, Givskov M, Kjelleberg S. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J Bacteriol. 2004;186:692–8. doi: 10.1128/JB.186.3.692-698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLean RJC, Whiteley M, Stickler DJ, Fuqua WC. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol Lett. 1997;154:259–63. doi: 10.1111/j.1574-6968.1997.tb12653.x. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–75. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanzelka BL, Greenberg EP. Quorum sensing in Vibrio fischeri: evidence that S-adenosylmethionine is the amino acid substrate for autoinducer synthesis. J Bacteriol. 1996;178:5291–4. doi: 10.1128/jb.178.17.5291-5294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson WT, Minogue TD, Val DL, von Bodman SB, Churchill ME. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol Cell. 2002;9:685–94. doi: 10.1016/S1097-2765(02)00480-X. [DOI] [PubMed] [Google Scholar]
- 12.Hanzelka BL, Stevens AM, Parsek MR, Crone TJ, Greenberg EP. Mutational analysis of the Vibrio fischeri LuxI polypeptide: critical regions of an autoinducer synthase. J Bacteriol. 1997;179:4882–7. doi: 10.1128/jb.179.15.4882-4887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullinane M, Baysse C, Morrissey JP, O’Gara F. Identification of two lysophosphatidic acid acyltransferase genes with overlapping function in Pseudomonas fluorescens. Microbiology. 2005;151:3071–80. doi: 10.1099/mic.0.27958-0. [DOI] [PubMed] [Google Scholar]
- 14.Beck von Bodman S, Farrand SK. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J Bacteriol. 1995;177:5000–8. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juhas M, Wiehlmann L, Salunkhe P, Lauber J, Buer J, Tümmler B. GeneChip expression analysis of the VqsR regulon of Pseudomonas aeruginosa TB. FEMS Microbiol Lett. 2005;242:287–95. doi: 10.1016/j.femsle.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–9. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piper KR, Beck von Bodman S, Farrand SK. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–50. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 18.d’Angelo-Picard C, Faure D, Penot I, Dessaux Y. Diversity of N-acyl homoserine lactone-producing and -degrading bacteria in soil and tobacco rhizosphere. Environ Microbiol. 2005;7:1796–808. doi: 10.1111/j.1462-2920.2005.00886.x. [DOI] [PubMed] [Google Scholar]
- 19.Dunphy G, Miyamoto C, Meighen E. A homoserine lactone autoinducer regulates virulence of an insect-pathogenic bacterium, Xenorhabdus nematophilus (Enterobacteriaceae) J Bacteriol. 1997;179:5288–91. doi: 10.1128/jb.179.17.5288-5291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinto UM, Viana ED, Martins ML, Vanetti MCD. Detection of acylated homoserine lactones in gram-negative proteolytic psychrotrophic bacteria isolated from cooled raw milk. Food Contr. 2007;18:1322–7. doi: 10.1016/j.foodcont.2006.09.005. [DOI] [Google Scholar]
- 21.Chong G, Kimyon O, Rice SA, Kjelleberg S, Manefield M. The presence and role of bacterial quorum sensing in activated sludge. Microb Biotechnol. 2012;5:621–33. doi: 10.1111/j.1751-7915.2012.00348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyer M, Wisniewski-Dyé F. Cell-cell signalling in bacteria: not simply a matter of quorum. FEMS Microbiol Ecol. 2009;70:1–19. doi: 10.1111/j.1574-6941.2009.00745.x. [DOI] [PubMed] [Google Scholar]
- 23.Eberl L, Riedel K. Mining quorum sensing regulated proteins - Role of bacterial cell-to-cell communication in global gene regulation as assessed by proteomics. Proteomics. 2011;11:3070–85. doi: 10.1002/pmic.201000814. [DOI] [PubMed] [Google Scholar]
- 24.Nakazawa H, Arakaki A, Narita-Yamada S, Yashiro I, Jinno K, Aoki N, Tsuruyama A, Okamura Y, Tanikawa S, Fujita N, et al. Whole genome sequence of Desulfovibrio magneticus strain RS-1 revealed common gene clusters in magnetotactic bacteria. Genome Res. 2009;19:1801–8. doi: 10.1101/gr.088906.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shelobolina ES, Vrionis HA, Findlay RH, Lovley DR. Geobacter uraniireducens sp. nov., isolated from subsurface sediment undergoing uranium bioremediation. Int J Syst Evol Microbiol. 2008;58:1075–8. doi: 10.1099/ijs.0.65377-0. [DOI] [PubMed] [Google Scholar]
- 26.Simmons SL, Dibartolo G, Denef VJ, Goltsman DS, Thelen MP, Banfield JF. Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS Biol. 2008;6:e177. doi: 10.1371/journal.pbio.0060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–94. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 28.Zhang G, Zhang F, Ding G, Li J, Guo X, Zhu J, Zhou L, Cai S, Liu X, Luo Y, et al. Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 2012;6:1336–44. doi: 10.1038/ismej.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray KM, Garey JR. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology. 2001;147:2379–87. doi: 10.1099/00221287-147-8-2379. [DOI] [PubMed] [Google Scholar]
- 30.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–69. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellenberger E. Exploring the unknown. The silent revolution of microbiology. EMBO Rep. 2001;2:5–7. doi: 10.1093/embo-reports/kve014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245–9. doi: 10.1016/S1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 33.Kimura N. Metagenomics: Access to unculturable microbes in the environment. Microbes Environ. 2006;21:201–15. doi: 10.1264/jsme2.21.201. [DOI] [Google Scholar]
- 34.Kimura N, Sakai K, Nakamura K. Isolation and characterization of a 4-nitrotoluene-oxidizing enzyme from activated sludge by a metagenomic approach. Microbes Environ. 2010;25:133–9. doi: 10.1264/jsme2.ME10110. [DOI] [PubMed] [Google Scholar]
- 35.Fujita MJ, Kimura N, Sakai A, Ichikawa Y, Hanyu T, Otsuka M. Cloning and heterologous expression of the vibrioferrin biosynthetic gene cluster from a marine metagenomic library. Biosci Biotechnol Biochem. 2011;75:2283–7. doi: 10.1271/bbb.110379. [DOI] [PubMed] [Google Scholar]
- 36.Fujita MJ, Kimura N, Yokose H, Otsuka M. Heterologous production of bisucaberin using a biosynthetic gene cluster cloned from a deep sea metagenome. Mol Biosyst. 2012;8:482–5. doi: 10.1039/c1mb05431g. [DOI] [PubMed] [Google Scholar]
- 37.Streit WR, Daniel R, Jaeger KE. Prospecting for biocatalysts and drugs in the genomes of non-cultured microorganisms. Curr Opin Biotechnol. 2004;15:285–90. doi: 10.1016/j.copbio.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 38.Schmeisser C, Steele H, Streit WR. Metagenomics, biotechnology with non-culturable microbes. Appl Microbiol Biotechnol. 2007;75:955–62. doi: 10.1007/s00253-007-0945-5. [DOI] [PubMed] [Google Scholar]
- 39.Kimura N, Sakai K, Nakamura K. Isolation and characterization of a 4-nitrotoluene-oxidizing enzyme from activated sludge by a metagenomic approach. Microbes Environ. 2010;25:133–9. doi: 10.1264/jsme2.ME10110. [DOI] [PubMed] [Google Scholar]
- 40.Craig JW, Chang FY, Kim JH, Obiajulu SC, Brady SF. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl Environ Microbiol. 2010;76:1633–41. doi: 10.1128/AEM.02169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williamson LL, Borlee BR, Schloss PD, Guan C, Allen HK, Handelsman J. Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl Environ Microbiol. 2005;71:6335–44. doi: 10.1128/AEM.71.10.6335-6344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao Y, Winans SC, Glick BR, Charles TC. Identification and characterization of new LuxR/LuxI-type quorum sensing systems from metagenomic libraries. Environ Microbiol. 2010;12:105–17. doi: 10.1111/j.1462-2920.2009.02049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–85. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan C, Ju J, Borlee BR, Williamson LL, Shen B, Raffa KF, Handelsman J. Signal mimics derived from a metagenomic analysis of the gypsy moth gut microbiota. Appl Environ Microbiol. 2007;73:3669–76. doi: 10.1128/AEM.02617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valle A, Bailey MJ, Whiteley AS, Manefield M. N-acyl-l-homoserine lactones (AHLs) affect microbial community composition and function in activated sludge. Environ Microbiol. 2004;6:424–33. doi: 10.1111/j.1462-2920.2004.00581.x. [DOI] [PubMed] [Google Scholar]
- 46.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–76. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 47.Onaka H, Ando N, Nihira T, Yamada Y, Beppu T, Horinouchi S. Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J Bacteriol. 1995;177:6083–92. doi: 10.1128/jb.177.21.6083-6092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, et al. A new class of homoserine lactone quorum-sensing signals. Nature. 2008;454:595–9. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- 49.Wang D, Ding X, Rather PN. Indole can act as an extracellular signal in Escherichia coli. J Bacteriol. 2001;183:4210–6. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anyanful A, Dolan-Livengood JM, Lewis T, Sheth S, Dezalia MN, Sherman MA, Kalman LV, Benian GM, Kalman D. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol Microbiol. 2005;57:988–1007. doi: 10.1111/j.1365-2958.2005.04739.x. [DOI] [PubMed] [Google Scholar]
- 51.Lee J, Attila C, Cirillo SLG, Cirillo JD, Wood TK. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb Biotechnol. 2009;2:75–90. doi: 10.1111/j.1751-7915.2008.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirakawa H, Kodama T, Takumi-Kobayashi A, Honda T, Yamaguchi A. Secreted indole serves as a signal for expression of type III secretion system translocators in enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 2009;155:541–50. doi: 10.1099/mic.0.020420-0. [DOI] [PubMed] [Google Scholar]
- 53.Mueller RS, Beyhan S, Saini SG, Yildiz FH, Bartlett DH. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J Bacteriol. 2009;191:3504–16. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martino PD, Fursy R, Bret L, Sundararaju B, Phillips RS. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol. 2003;49:443–9. doi: 10.1139/w03-056. [DOI] [PubMed] [Google Scholar]
- 55.Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindum PW, Anthoni U, Christophersen C, Eberl L, Molin S, Givskov M. N-Acyl-L-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:6384–8. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu L, Li H, Vuong C, Vadyvaloo V, Wang J, Yao Y, Otto M, Gao Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2006;74:488–96. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burr T, Barnard AM, Corbett MJ, Pemberton CL, Simpson NJ, Salmond GP. Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol Microbiol. 2006;59:113–25. doi: 10.1111/j.1365-2958.2005.04939.x. [DOI] [PubMed] [Google Scholar]
- 59.Givskov M, Ostling J, Eberl L, Lindum PW, Christensen AB, Christiansen G, Molin S, Kjelleberg S. Two separate regulatory systems participate in control of swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:742–5. doi: 10.1128/jb.180.3.742-745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Labbate M, Queck SY, Koh KS, Rice SA, Givskov M, Kjelleberg S. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J Bacteriol. 2004;186:692–8. doi: 10.1128/JB.186.3.692-698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–4. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 62.Leadbetter JR, Greenberg EP. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J Bacteriol. 2000;182:6921–6. doi: 10.1128/JB.182.24.6921-6926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang JJ, Han JI, Zhang LH, Leadbetter JR. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2003;69:5941–9. doi: 10.1128/AEM.69.10.5941-5949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu JY, Fan Y, Lin YH, Zhang HB, Ong SL, Dong N, Xu JL, Ng WJ, Zhang LH. Microbial diversity and prevalence of virulent pathogens in biofilms developed in a water reclamation system. Res Microbiol. 2003;154:623–9. doi: 10.1016/j.resmic.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 65.Flagan S, Ching WK, Leadbetter JR. Arthrobacter strain VAI-A utilizes acyl-homoserine lactone inactivation products and stimulates quorum signal biodegradation by Variovorax paradoxus. Appl Environ Microbiol. 2003;69:909–16. doi: 10.1128/AEM.69.2.909-916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uroz S, D’Angelo-Picard C, Carlier A, Elasri M, Sicot C, Petit A, Oger P, Faure D, Dessaux Y. Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology. 2003;149:1981–9. doi: 10.1099/mic.0.26375-0. [DOI] [PubMed] [Google Scholar]
- 67.Lin YH, Xu JL, Hu J, Wang LH, Ong SL, Leadbetter JR, Zhang LH. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol Microbiol. 2003;47:849–60. doi: 10.1046/j.1365-2958.2003.03351.x. [DOI] [PubMed] [Google Scholar]
- 68.Park SY, Lee SJ, Oh TK, Oh JW, Koo BT, Yum DY, Lee JK. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology. 2003;149:1541–50. doi: 10.1099/mic.0.26269-0. [DOI] [PubMed] [Google Scholar]
- 69.Yoon JH, Lee JK, Jung SY, Kim JA, Kim HK, Oh TK. Nocardioides kongjuensis sp. nov., an N-acylhomoserine lactone-degrading bacterium. Int J Syst Evol Microbiol. 2006;56:1783–7. doi: 10.1099/ijs.0.64120-0. [DOI] [PubMed] [Google Scholar]
- 70.Romero M, Avendaño-Herrera R, Magariños B, Cámara M, Otero A. Acylhomoserine lactone production and degradation by the fish pathogen Tenacibaculum maritimum, a member of the Cytophaga-Flavobacterium-Bacteroides (CFB) group. FEMS Microbiol Lett. 2010;304:131–9. doi: 10.1111/j.1574-6968.2009.01889.x. [DOI] [PubMed] [Google Scholar]
- 71.Romero M, Diggle SP, Heeb S, Cámara M, Otero A. Quorum quenching activity in Anabaena sp. PCC 7120: identification of AiiC, a novel AHL-acylase. FEMS Microbiol Lett. 2008;280:73–80. doi: 10.1111/j.1574-6968.2007.01046.x. [DOI] [PubMed] [Google Scholar]
- 72.Huang JJ, Han JI, Zhang LH, Leadbetter JR. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2003;69:5941–9. doi: 10.1128/AEM.69.10.5941-5949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sio CF, Otten LG, Cool RH, Diggle SP, Braun PG, Bos R, Daykin M, Cámara M, Williams P, Quax WJ. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun. 2006;74:1673–82. doi: 10.1128/IAI.74.3.1673-1682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dong YH, Xu JL, Li XZ, Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci U S A. 2000;97:3526–31. doi: 10.1073/pnas.97.7.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee SJ, Park SY, Lee JJ, Yum DY, Koo BT, Lee JK. Genes encoding the N-acyl homoserine lactone-degrading enzyme are widespread in many subspecies of Bacillus thuringiensis. Appl Environ Microbiol. 2002;68:3919–24. doi: 10.1128/AEM.68.8.3919-3924.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang WZ, Morohoshi T, Ikenoya M, Someya N, Ikeda T. AiiM, a novel class of N-acylhomoserine lactonase from the leaf-associated bacterium Microbacterium testaceum. Appl Environ Microbiol. 2010;76:2524–30. doi: 10.1128/AEM.02738-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park SY, Hwang BJ, Shin MH, Kim JA, Kim HK, Lee JK. N-acylhomoserine lactonase producing Rhodococcus spp. with different AHL-degrading activities. FEMS Microbiol Lett. 2006;261:102–8. doi: 10.1111/j.1574-6968.2006.00336.x. [DOI] [PubMed] [Google Scholar]
- 78.Park SY, Kang HO, Jang HS, Lee JK, Koo BT, Yum DY. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl Environ Microbiol. 2005;71:2632–41. doi: 10.1128/AEM.71.5.2632-2641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uroz S, Oger PM, Chapelle E, Adeline MT, Faure D, Dessaux Y. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl Environ Microbiol. 2008;74:1357–66. doi: 10.1128/AEM.02014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uroz S, Chhabra SR, Cámara M, Williams P, Oger P, Dessaux Y. N-Acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology. 2005;151:3313–22. doi: 10.1099/mic.0.27961-0. [DOI] [PubMed] [Google Scholar]
- 81.Zelinski T, Kula MR. A kinetic study and application of a novel carbonyl reductase isolated from Rhodococcus erythropolis. Bioorg Med Chem. 1994;2:421–8. doi: 10.1016/0968-0896(94)80010-3. [DOI] [PubMed] [Google Scholar]
- 82.Zelinski T, Peters J, Kula MR. Purification and characterization of a novel carbonyl reductase isolated from Rhodococcus erythropolis. J Biotechnol. 1994;33:283–92. doi: 10.1016/0168-1656(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 83.Kalia VC, Raju SC, Purohit HJ. Genomic analysis reveals versatile organisms for quorum quenching enzymes: acyl-homoserine lactone-acylase and -lactonase. Open Microbiol J. 2011;5:1–13. doi: 10.2174/1874285801105010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Riaz K, Elmerich C, Moreira D, Raffoux A, Dessaux Y, Faure D. A metagenomic analysis of soil bacteria extends the diversity of quorum-quenching lactonases. Environ Microbiol. 2008;10:560–70. doi: 10.1111/j.1462-2920.2007.01475.x. [DOI] [PubMed] [Google Scholar]
- 85.Tannières M, Beury-Cirou A, Vigouroux A, Mondy S, Pellissier F, Dessaux Y, Faure D. A metagenomic study highlights phylogenetic proximity of quorum-quenching and xenobiotic-degrading amidases of the AS-family. PLoS One. 2013;8:e65473. doi: 10.1371/journal.pone.0065473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Christensen QH, Grove TL, Booker SJ, Greenberg EP. A high-throughput screen for quorum-sensing inhibitors that target acyl-homoserine lactone synthases. Proc Natl Acad Sci U S A. 2013;110:13815–20. doi: 10.1073/pnas.1313098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Müh U, Schuster M, Heim R, Singh A, Olson ER, Greenberg EP. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob Agents Chemother. 2006;50:3674–9. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ni N, Li M, Wang J, Wang B. Inhibitors and antagonists of bacterial quorum sensing. Med Res Rev. 2009;29:65–124. doi: 10.1002/med.20145. [DOI] [PubMed] [Google Scholar]
- 89.Chen G, Swem LR, Swem DL, Stauff DL, O’Loughlin CT, Jeffrey PD, Bassler BL, Hughson FM. A strategy for antagonizing quorum sensing. Mol Cell. 2011;42:199–209. doi: 10.1016/j.molcel.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl Environ Microbiol. 2007;73:4100–9. doi: 10.1128/AEM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tashiro Y, Toyofuku M, Nakajima-Kambe T, Uchiyama H, Nomura N. Bicyclic compounds repress membrane vesicle production and Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2010;304:123–30. doi: 10.1111/j.1574-6968.2010.01897.x. [DOI] [PubMed] [Google Scholar]
- 92.Ren D, Bedzyk LA, Ye RW, Thomas SM, Wood TK. Stationary-phase quorum-sensing signals affect autoinducer-2 and gene expression in Escherichia coli. Appl Environ Microbiol. 2004;70:2038–43. doi: 10.1128/AEM.70.4.2038-2043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Henne A, Daniel R, Schmitz RA, Gottschalk G. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl Environ Microbiol. 1999;65:3901–7. doi: 10.1128/aem.65.9.3901-3907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lämmle K, Zipper H, Breuer M, Hauer B, Buta C, Brunner H, Rupp S. Identification of novel enzymes with different hydrolytic activities by metagenome expression cloning. J Biotechnol. 2007;127:575–92. doi: 10.1016/j.jbiotec.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 95.Courtois S, Cappellano CM, Ball M, Francou FX, Normand P, Helynck G, Martinez A, Kolvek SJ, Hopke J, Osburne MS, et al. Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl Environ Microbiol. 2003;69:49–55. doi: 10.1128/AEM.69.1.49-55.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Entcheva P, Liebl W, Johann A, Hartsch T, Streit WR. Direct cloning from enrichment cultures, a reliable strategy for isolation of complete operons and genes from microbial consortia. Appl Environ Microbiol. 2001;67:89–99. doi: 10.1128/AEM.67.1.89-99.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knietsch A, Waschkowitz T, Bowien S, Henne A, Daniel R. Construction and screening of metagenomic libraries derived from enrichment cultures: generation of a gene bank for genes conferring alcohol oxidoreductase activity on Escherichia coli. Appl Environ Microbiol. 2003;69:1408–16. doi: 10.1128/AEM.69.3.1408-1416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Voget S, Leggewie C, Uesbeck A, Raasch C, Jaeger KE, Streit WR. Prospecting for novel biocatalysts in a soil metagenome. Appl Environ Microbiol. 2003;69:6235–42. doi: 10.1128/AEM.69.10.6235-6242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nasuno E, Kimura N, Fujita MJ, Nakatsu CH, Kamagata Y, Hanada S. Phylogenetically novel LuxI/LuxR-type quorum sensing systems isolated using a metagenomic approach. Appl Environ Microbiol. 2012;78:8067–74. doi: 10.1128/AEM.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Suenaga H, Ohnuki T, Miyazaki K. Functional screening of a metagenomic library for genes involved in microbial degradation of aromatic compounds. Environ Microbiol. 2007;9:2289–97. doi: 10.1111/j.1462-2920.2007.01342.x. [DOI] [PubMed] [Google Scholar]
- 101.Gloux K, Leclerc M, Iliozer H, L’Haridon R, Manichanh C, Corthier G, Nalin R, Blottière HM, Doré J. Development of high-throughput phenotyping of metagenomic clones from the human gut microbiome for modulation of eukaryotic cell growth. Appl Environ Microbiol. 2007;73:3734–7. doi: 10.1128/AEM.02204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wynendaele E, Bronselaer A, Nielandt J, D’Hondt M, Stalmans S, Bracke N, Verbeke F, Van De Wiele C, De Tré G, De Spiegeleer B. Quorumpeps database: chemical space, microbial origin and functionality of quorum sensing peptides. Nucleic Acids Res. 2013;41:D655–9. doi: 10.1093/nar/gks1137. [DOI] [PMC free article] [PubMed] [Google Scholar]