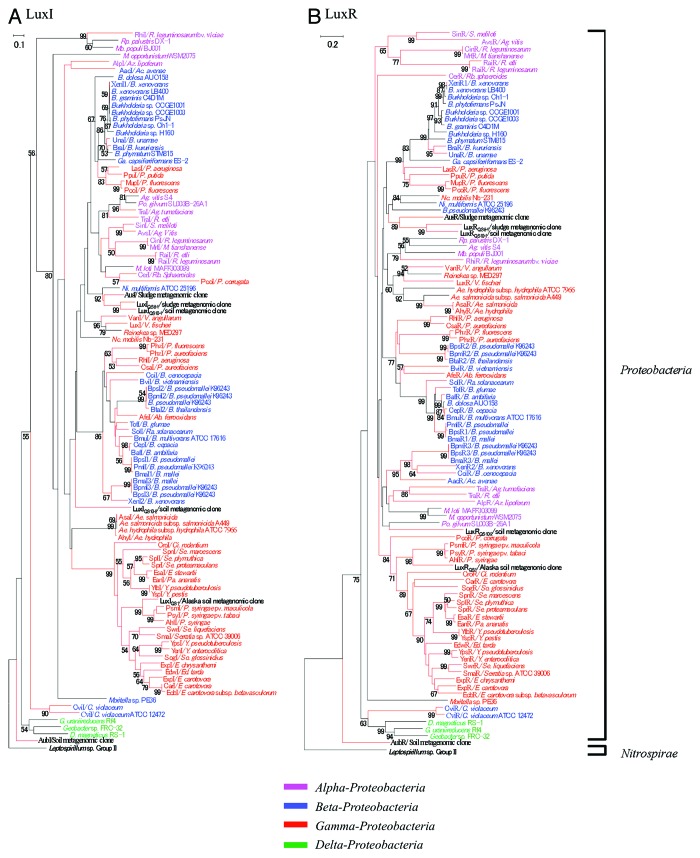
Figure 3. Phylogenetic trees of (A) LuxI and (B) LuxR protein sequence homologs obtained from Proteobacteria and Nitrospirae. The members isolated from metagenomic clones are indicated in bold type. The characterized proteins AubI, AubR, AusI, and AusR are highlighted by red closed boxes. Species belonging to the α-, β-, γ-, and δ-Proteobacteria are grouped within brackets. A Leptospirillum sequence is used as an outgroup. The bold brackets represent two groups of phyla. The branch length and bootstrap values were obtained from 1000 replicates using a neighbor-joining algorithm. Nodes lacking a number are those that could not be supported by more than 50% of the bootstrap value. The scale bars represent 0.1 and 0.2 substitutions per amino acid site, respectively. The red branches are sequences with experimental evidence. Abbreviations for the bacterial genus names are as follows: Ab, Acidithiobacillus; Ac, Acidovorax; Ae, Aeromonas; Ag, Agrobacterium; Az, Azospirillum; B, Burkholderia; C, Chromobacterium; Ci, Citrobacter; D, Desulfovibrio; E, Erwinia; Ed, Edward; G, Geobacter; Ga, Gallionella; M, Mesorhizobium; Mb, Methylobacterium; Ni, Nitrosospira; Nc, Nitrococcus; P, Pseudomonas; Pa, Pantoea; Po, Polymorphum; R, Rhizobium; Ra, Ralstonia; Rb, Rhodobacter; Rp, Rhodopseudomonas; S, Sinorhizobium; Se, Serratia; So, Sodalis; V, Vibrio; Y, Yersinia.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.