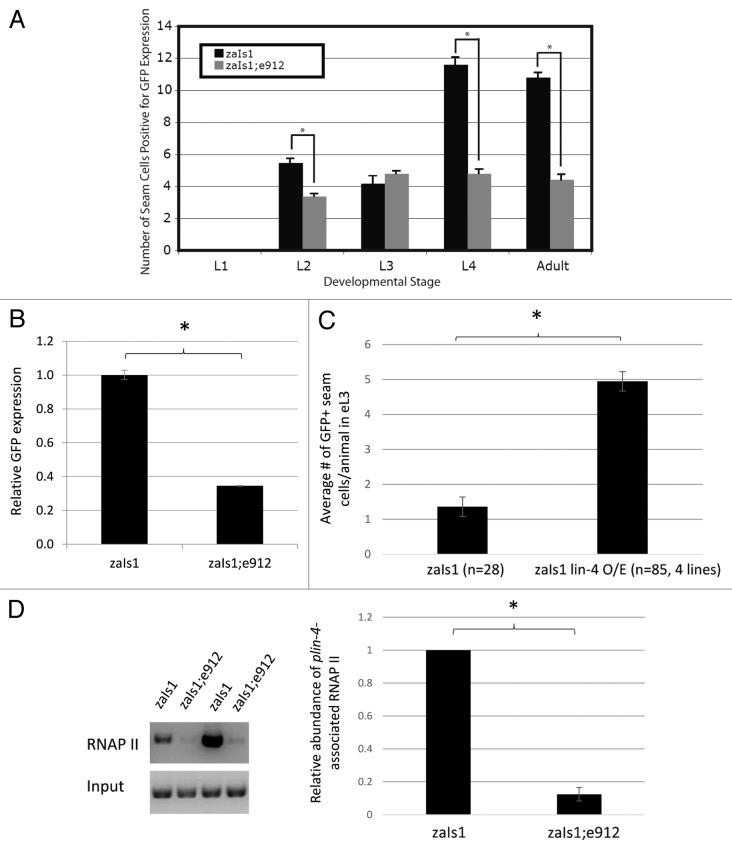
Figure 3.lin-4 is necessary and sufficient for transcriptional autoactivation. (A) GFP expression data in animals carrying the lin-4:GFP integrated transgene in the wild-type (zaIs1) and the lin-4 (e912) deletion background. Seam cells positive for GFP were counted at each larval stage of development and in adulthood. Error bars represent the standard error of the mean, and significance was measured using a 2-sample t test; P values are as indicated (*P < 1 × 10−6); n = 50, zaIs1; n = 150, zaIs1;e912. (B) Whole-body GFP mRNA levels collected from mixed-stage animals were measured by qPCR and represented as the mean ± SEM. Expression levels were normalized to pmp-3 mRNA. Significance was measured using a 2-tailed t test (*P < 0.0001). (C) GFP+ seam cell counts in synchronized, early L3 (eL3) animals, represented as the mean ± SEM. GFP+ seam cell counts for the lin-4 overexpressor lines (O/E) are an average from 4 independent transgenic lines. Significance was measured using a 2-tailed t test (*P < 1 × 10−10). (D) RNAP II chromatin immunoprecipitation from zaIs1 and zaIs1;e912 mixed-stage populations grown in liquid culture (see methods). Left panel: semi-quantitative PCR detecting a ~500 bp fragment of the lin-4 promoter upstream of GFP, from 2 technical ChIP replicates. Right panel: ImageJ quantification of band intensities. The data shown is an average of the 2 replicates from the gel in the left panel and an additional biological replicate (not shown in gel). Significance was measured using a 2-tailed t test (*P = 0.002). Error bars represent standard deviation of the mean.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.