Abstract
Although the roles of mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3 kinase (PI3K) signaling in KRAS-driven tumorigenesis are well established, KRAS activates additional pathways required for tumor maintenance, inhibition of which are likely to be necessary for effective KRAS-directed therapy. Here we show that the IKK-related kinases TBK1 and IKKε promote KRAS-driven tumorigenesis by regulating autocrine CCL5 and IL-6 and identify CYT387 as a potent JAK/TBK1/IKKε inhibitor. CYT387 treatment ablates RAS-associated cytokine signaling and impairs Kras-driven murine lung cancer growth. Combined CYT387 and MEK inhibitor therapy induces regression of aggressive murine lung adenocarcinomas driven by Kras mutation and p53 loss. These observations reveal that TBK1/IKKε promote tumor survival by activating CCL5 and IL-6 and identify concurrent inhibition of TBK1/IKKε, JAK, and MEK signaling as an effective approach to inhibit the actions of oncogenic KRAS.
Keywords: KRAS, cytokines, TBK1/IKKε, JAK, lung cancer
INTRODUCTION
Oncogenic mutations in KRAS and receptor tyrosine kinases (RTKs) drive tumor growth by engaging multiple downstream mitogenic pathways, including RAF-MAPK and PI3K-AKT (1). KRAS also activates RAL-GEF (2, 3) and inflammatory signals such as NF-κB (4–6). Although concurrent MAPK/PI3K pathway inhibition is under clinical evaluation, multiple approaches are likely necessary to identify effective KRAS targeted therapy.
Oncogenic RAS induces inflammatory cytokines that activate NF-κB and STAT3. For example, KRAS-driven non-small cell lung cancers (NSCLC) (7) and pancreatic ductal adenocarcinomas (PDAC) (8) engage cell autonomous IL-1 signaling. Similarly, oncogenic RAS induced IL-6 promotes both oncogene-induced senescence (9) and cell transformation (10), and STAT3 is required for Kras-driven PDAC development in mice (11–13). RAS-associated IL-8, CXCL1, CXCL2 expression also promotes senescence (14), tumor cell survival and angiogenesis (15, 16). How oncogenic RAS activates these cytokines and their role in RAS-dependent cancers remains incompletely characterized.
Activation of RALA and RALB by RAL-GEF enhances cancer cell proliferation and survival (17). A specific RALB-SEC5 complex engages the innate immune signaling kinase Tank Binding Kinase-1 (TBK1) to promote cell survival (18). TBK1 is required for transformation by oncogenic KRAS, sustains KRAS-dependent cancer cell viability, and regulates basal autophagy (18–22). The TBK1 homologue IKKε (encoded by IKBKE) also promotes NF-κB activation downstream of KRAS (23), substitutes for AKT to drive cell transformation (24), and is induced by RAS-associated cytokines such as IL-1 and IL-6 (25). Thus, TBK1/IKKε signaling is coopted by oncogenic KRAS and facilitates tumorigenesis.
Following viral infection, TBK1 and IKKε amplify IFN-β production via an autocrine loop (26). Here we identify a similar circuit involving CCL5 and IL-6 required for KRAS-driven lung tumorigenesis and potently suppressed by CYT387, a novel TBK1/IKKε and JAK inhibitor.
RESULTS
TBK1 regulated cell survival involves autocrine CCL5 and IL-6 and STAT3 signaling
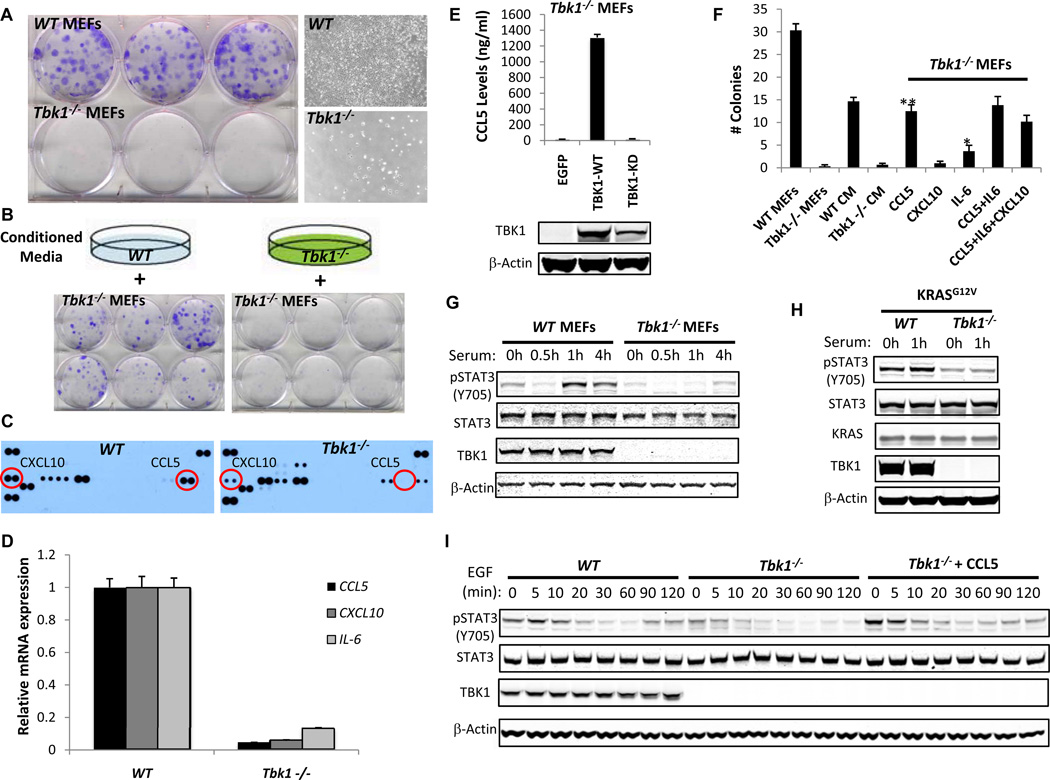
Expression of Tbk1 is required for transformation by oncogenic KRAS (18, 20, 21). Although Tbk1−/− MEFs proliferate in standard culture, we noted marked impairment of Tbk1−/− MEF proliferation in a clonogenic assay compared with WT littermate control MEFs (Fig. 1A). To assess the role of cell contact versus a secreted factor, we plated Tbk1−/− MEFs clonogenically in conditioned medium (CM) from WT or Tbk1−/− MEFs propagated at high density (Fig. 1B). CM from WT but not Tbk1−/− MEFs rescued colony formation, revealing that Tbk1 regulates secreted factors that promote cell proliferation and may contribute to KRAS-driven tumorigenesis.
Figure 1. Autocrine CCL5 drives TBK1-dependent clonogenic proliferation.
A, Crystal violet stain and phase-contrast images (10×) of WT or Tbk1−/− MEFs cultured in a clonogenic assay for 10 d.
B, Crystal violet stain of Tbk1−/− MEFs cultured clonogenically in CM from WT or Tbk1−/− MEFs.
C, Cytokine antibody arrays of CM from WT or Tbk1−/− MEFs.
D, mRNA levels of CCL5, CXCL10, or IL-6 in WT or Tbk1−/− MEFs. Relative expression normalized to WT MEF mRNA levels, mean and SEM of triplicate samples shown. p < 0.001 for each comparison by t-test.
E, CCL5 levels by ELISA 72 h after EGFP, TBK1-WT, or TBK1-KD re-expression in Tbk1−/− MEFs. Mean ± SD of duplicate samples shown. TBK1 and β-actin immunoblot below.
F, Colony number following media supplementation of Tbk1−/− MEFs, mean ± SEM of 6 replicates shown. **p<0.001 and *p<0.01 for comparison to untreated Tbk1−/− MEFs by t-test.
G, Immunoblot of Y705 pSTAT3, STAT3, TBK1 and β-Actin levels in WT or Tbk1−/− MEFs serum-starved overnight and stimulated with 10% serum for the indicated times.
H, Immunoblot of Y705 pSTAT3, STAT3, TBK1, KRAS and β-Actin levels 48 h following KRASG12V expression in serum-starved and stimulated WT or Tbk1−/− MEFs.
I, WT or Tbk1−/− MEFs ± CCL5 (10 ng/ml) stimulated with EGF (100 ng/ml). Immunoblot shows Y705 pSTAT3, STAT3, TBK1, and β-Actin.
Since TBK1/IKKε regulate cytokine production, we assessed CM from WT or Tbk1−/− MEFs using a cytokine antibody array. Tbk1−/− MEF CM lacked CCL5 and exhibited decreased CXCL10 levels compared to WT MEF CM (Fig. 1C and Supplementary Fig. S1A). CCL5 and CXCL10 were also absent in Tbk1−/− MEF clonogenic culture media (Supplementary Fig. S1B). Since Tbk1 also regulates IL-6 (27), we measured CCL5, CXCL10 and IL-6 mRNA levels and observed reduced expression of each cytokine/chemokine in Tbk1−/− MEFs (Fig. 1D), whereas others such as CXCL1 were increased (Supplementary Fig. S1C and S1D). Re-introduction of WT but not kinase dead (KD) TBK1 restored CCL5 production by Tbk1−/− MEFs, revealing kinase-dependent regulation (Fig. 1E).
To examine the contribution of CCL5, CXCL10, and/or IL-6 to TBK1-regulated survival, we supplemented media with each factor and measured Tbk1−/− MEF colony formation. CCL5 (10 ng/ml) rescued Tbk1−/− MEFs colonies comparably with WT MEF CM, whereas IL-6 had a modest effect and CXCL10 failed to rescue colony formation (Fig. 1F and Supplementary Fig. S1E). Adding IL-6 or CXCL10 to CCL5 did not increase Tbk1−/− MEF colonies. Thus, autocrine CCL5 and IL-6 signaling promote TBK1-regulated proliferation/survival.
Since CCL5 and IL-6 induce pro-survival JAK-STAT signaling, we next measured Y705 pSTAT3 phosphorylation in WT or Tbk1−/− MEFs. Tbk1−/− MEFs exhibited low pSTAT3 levels following release from serum starvation (Fig. 1G). Oncogenic KRASG12V expression or stimulation of RAS activity with EGF failed to rescue STAT3 signaling in Tbk1−/− MEFs (Fig. 1H and I). Supplementation of Tbk1−/− MEF media with CCL5 completely restored pSTAT3 activation under basal conditions and following EGF stimulation (Fig. 1I). Thus, TBK1-regulated CCL5 promotes both clonogenic proliferation and autocrine STAT3 activation.
CYT387 is a potent TBK1/IKKε inhibitor
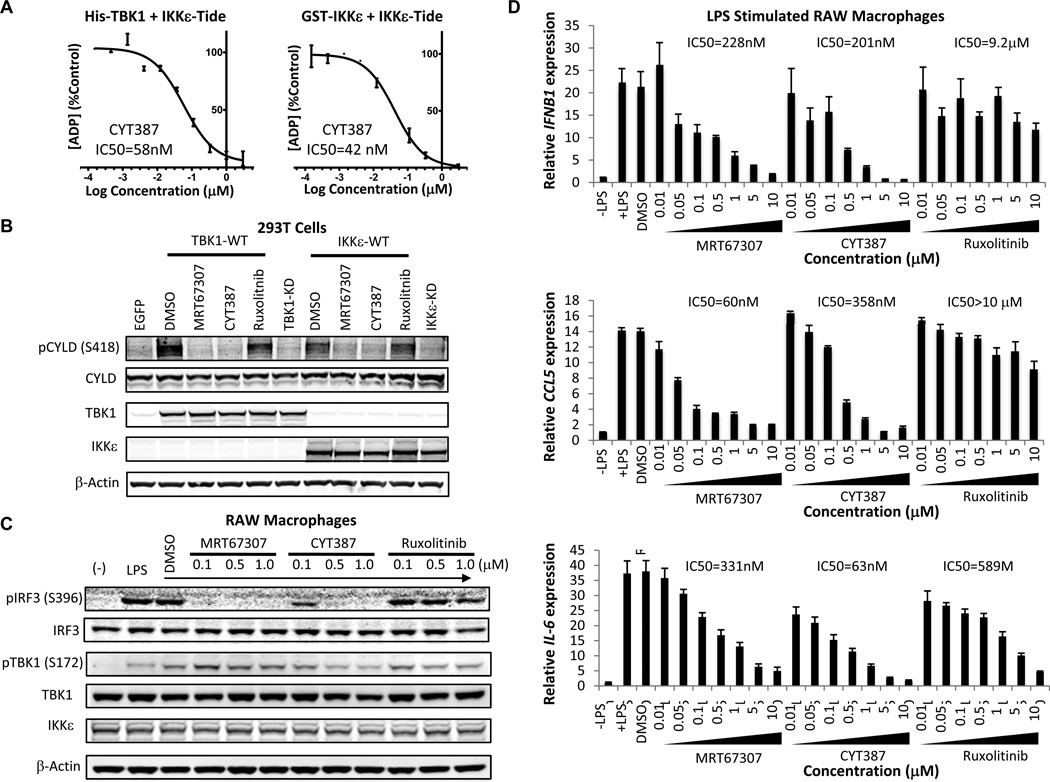
MRT67307 has been characterized as a potent TBK1 inhibitor (28). We noted that CYT387, a JAK1/2 kinase inhibitor in clinical trials for myelofibrosis, also inhibited TBK1 with low nanomolar potency (29). To explore the TBK1- and IKKε–specific activities of CYT387, we first measured TBK1/IKKε-mediated phosphorylation of a substrate motif peptide (IKKε-Tide) in vitro (30). CYT387 potently inhibited TBK1 (IC50 = 58 nM) and IKKε (IC50 = 42 nM) kinase activity in the presence of 0.1 mM ATP (Fig. 2A). In contrast, another JAK1/2 inhibitor, Ruxolitinib, failed to inhibit TBK1 or IKKε in this assay (IC50 > 1µM for both) (Supplementary Fig. S2A). MRT67307 was comparable to CYT387 in the TBK1 assay (IC50 = 40 nM), but inhibited IKKε less potently (IC50 = 242 nM) (Supplementary Fig. S2A). To confirm these observations in intact cells, we examined the effect of inhibitor treatment on TBK1/IKKε S418 CYLD phosphorylation, which mediates IKKε-induced transformation (30). Treatment with CYT387 abrogated TBK1/IKKε-induced CYLD phosphorylation in 293T cells, similar to MRT67307 and in contrast to Ruxolitinib (Fig. 2B). These findings established CYT387 as a potent TBK1/IKKε inhibitor.
Figure 2. CYT387 inhibits JAK and TBK1/IKKε signaling.
A, In vitro kinase assay with His-TBK1 (4 nM) or GST-IKKε (2 nM), 100 µM IKKε-tide, 100 µM ATP, and CYT387. Assays performed in duplicate, ADP generated normalized to DMSO control, mean ± SD shown.
B, Immunoblot of S418 pCYLD, CYLD, TBK1, IKKε, and β-Actin levels in 293T cells 24 h following transient co-transfection of Myc-tagged CYLD with TBK1-WT, TBK1-KD, IKKε-WT, or IKKε-KD and 4 h treatment with DMSO or 5 µM MRT67307, CYT387, or Ruxolitinib.
C, Immunoblot of S396 pIRF3, IRF3, S172 pTBK1, TBK1, IKKε, and β-Actin levels in macrophages stimulated with LPS (100 ng/ml) for 2 h ± DMSO or inhibitor pretreatment at the indicated concentrations for 1 h.
D, mRNA levels of IFNB1, CCL5, and IL6 in macrophages stimulated with LPS (100 ng/ml) for 2 h ± pretreatment with inhibitors for 1 h. Mean and SEM of triplicate samples shown.
To determine activity of these inhibitors in a physiological setting we next measured IFNγ-induced JAK activity or LPS-induced TBK1/IKKε signaling in murine RAW macrophages. As expected, Ruxolitinib treatment potently suppressed IFNγ-induced Y701 pSTAT1, in contrast to MRT67307 (Supplementary Fig. S2B). CYT387 was less potent than Ruxolitinib, but suppressed the STAT1 target gene GBP1 at higher concentrations (IC50 = 587) like Ruxolitinib (IC50 = 20 nM) and in contrast to MRT67307 (IC50 > 10 µM) (Supplementary Fig. S2C). Similar results were obtained for IFN-γ-induced CXCL10 mRNA expression (Supplementary Fig. S2D). CYT387 treatment potently inhibited LPS-induced S396 IRF3 phosphorylation at concentrations <1 µM, similar to MRT67307 and in contrast to Ruxolitinib (Fig. 2C). As previously reported MRT67307 treatment paradoxically induced TBK1 S172 activation loop phosphorylation (28), which was less pronounced following CYT387 treatment in this assay. MRT67307 (IC50 = 228 nM) or CYT387 (IC50 = 201 nM) treatment also suppressed expression of the IRF3 target gene IFNB1 (Fig. 2D). MRT67307 or CYT387 further impaired LPS-induced expression of CCL5 and CXCL10, whereas Ruxolitinib failed to suppress IFNB1, CCL5, or CXCL10 expression (Fig. 2D and Supplementary Fig. S2E). MRT67307 (IC50 = 331 nM) or Ruxolitinib (IC50 = 589 nM) each partially suppressed LPS-induced IL-6, whereas CYT387 strongly inhibited IL-6 mRNA levels (IC50 = 63 nM) (Fig. 2D). These findings confirmed CYT387 as a multitargeted JAK and TBK1/IKKε inhibitor (Supplementary Fig. S2F) that potently suppresses multiple cytokines, including CCL5 and IL-6.
CCL5 and IL-6 promote human KRAS-dependent lung cancer transformation
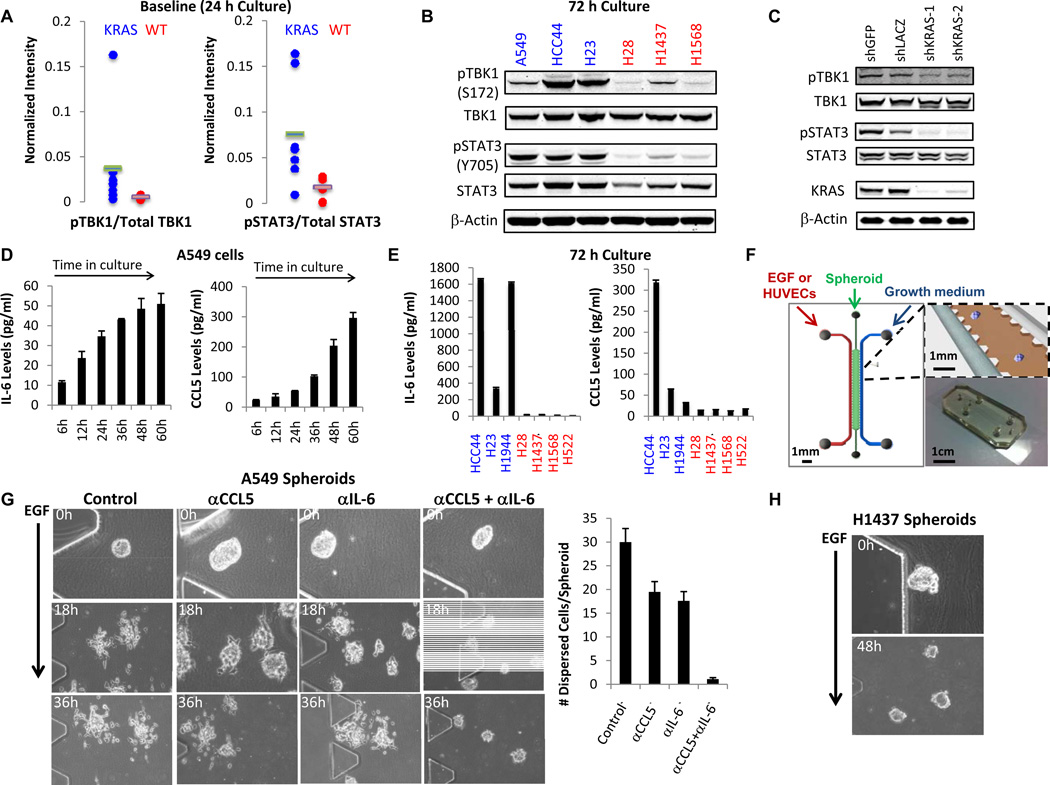
To determine the role of these cytokines in human KRAS-driven lung cancer, we first measured TBK1 and STAT3 activation in a panel of human NSCLC lines. Since KRAS dependency does not strictly correlate with KRAS mutation status (19, 20, 31), we examined specific cell lines for their dependence on KRAS expression (Supplementary Fig. S3A). Although one group reported that A549 cells are KRAS independent (31), we observed sensitivity of this cell line to KRAS and TBK1 suppression, consistent with other reports (19, 22, 32, 33). The majority of other KRAS dependent NSCLC cell lines also required TBK1 for survival (Supplementary Fig. S3A). We then compared activation of TBK1 and cytokine signaling in a panel of KRAS mutant/dependent cell lines (A549, HCC44, H23, H1792, H460, H2009, and H1944 cells) to KRAS WT/independent cells (Fig. 3A and Supplementary Fig. S3B) (19). We measured TBK1 activity by S172 phosphorylation and STAT3 activity by Y705 phosphorylation 24 h after plating, and normalized values to total TBK1 or STAT3 levels respectively. Although pTBK1 levels were low at baseline, KRAS-dependent NSCLC cells exhibited higher levels of both pTBK1 and pSTAT3 compared with KRAS-independent cells (Fig. 3A, Supplementary Fig S3B). Activation of pTBK1 and pSTAT3 was more pronounced in KRAS dependent NSCLC cells after 72 h in culture (Fig. 3B). Suppression of KRAS in A549 cells reduced levels of both pTBK1 and pSTAT3 (Fig. 3C), consistent with direct engagement of RALB-TBK1 signaling by oncogenic KRAS (18).
Figure 3. CCL5 and IL-6 promote RAS-driven lung cancer cell proliferation in 3D culture.
A, Baseline pTBK1/total TBK1 levels and pSTAT3/total STAT3 levels in NSCLC cell line panel 24 h after plating. Blue = KRAS mutant/dependent, Red = KRAS WT/independent.
B, Immunoblot of S172 pTBK1, TBK1, Y705 pSTAT3, STAT3, and β-Actin levels in A549, HCC44, H23, H28, H1437, or H1568 cells after 72 h culture. Blue = KRAS mutant/dependent, Red = KRAS WT/independent.
C, Immunoblot of pTBK1, TBK1, pSTAT3, STAT3, KRAS, and β-Actin levels in A549 cells 72 h following expression of control or KRAS-specific shRNAs.
D, IL-6 and CCL5 ELISA from media during A549 cell culture. Assays performed in duplicate, mean and SD shown.
E, IL-6 and CCL5 ELISA after 72 h in culture of additional KRAS dependent (blue) or independent (red) cell lines. Mean and SD of duplicate samples shown.
F, Schematic of microfluidic 3D cell culture device.
G, Phase-contrast images of A549 spheroids at 0 (20×), 18 and 36 h (10×) following EGF stimulation (20 ng/ml) ± neutralizing antibodies against CCL5 (100 ng/ml), IL-6 (100 ng/ml), or the combination. Mean ± SD number of dispersed cells per spheroid from triplicate devices shown.
H, Phase-contrast images H1437 spheroids 0 h (20×) or 48 h (10×) following EGF stimulation (20 ng/ml).
These findings suggested that autocrine cytokine signaling downstream of oncogenic KRAS contributes to the proliferation of KRAS-dependent cells. Indeed, A549 and other KRAS-dependent cell lines secreted CCL5 and IL-6 into the media over time in culture, in contrast to KRAS-independent cells (Fig. 3D and E). Extracellular signals in the tumor microenvironment such as IL-1 and EGF further enhanced TBK1 activation as well as CCL5 and IL-6 production in A549 cells (Supplementary Fig. S3C and D). To study the functional consequences of this autocrine cytokine production, we assessed CCL5 and IL-6 neutralization in 2D culture or in a microfluidic 3D cell culture system where A549 lung tumor spheroids are co-cultured in collagen with human vascular endothelial cells (HUVECs) or growth factors such as EGF (34) (Fig. 3F and Supplementary Fig. S3E). This 3D system more closely recapitulates cancer cell behavior in the extracellular matrix, captures features of the tumor microenvironment, and also enables inhibitor studies. Incubation of A549 cells with CCL5 and IL-6 neutralizing antibodies in 2D culture had a minor effect on proliferation (Supplementary Fig. S3F). In contrast, combined CCL5 and IL-6 blockade completely suppressed A549 cell proliferation in response to EGF in 3D culture, compared with neutralization of either cytokine alone (Fig. 3G). We also observed that KRAS-independent H1437 spheroids failed to disperse and proliferate in response to EGF even at 48 h (Fig. 3H). Together, these findings demonstrate that TBK1 signaling and CCL5 and IL-6 production not only promote clonogenic proliferation of MEFs, but also KRAS-dependent lung cancer cell proliferation and migration.
CYT387 treatment impairs KRAS-dependent NSCLC viability
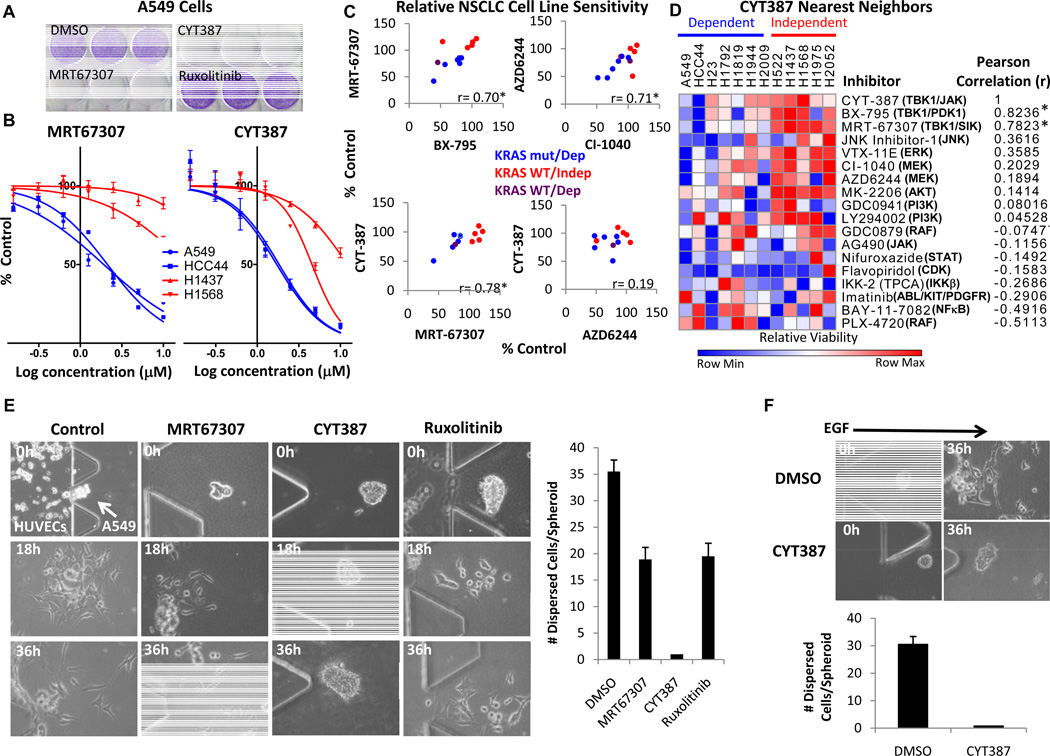
We next assessed the capacity of CYT387 to target this signaling pathway and impair the tumorigenic potential of KRAS-dependent lung cancer cells. Treatment of A549 cells with MRT67307 or CYT387 suppressed clonogenic growth and induced cytotoxicity, in contrast to Ruxolitinib (Fig. 4A and Supplementary Fig. S4A). Both MRT67307 and CYT387 preferentially impaired the viability of KRAS-dependent A549 and HCC44 cells (IC-50 ~ 2 µM) compared with KRAS-independent H1437 and H1568 cells, whereas Ruxolitinib had a negligible effect (Fig. 4B and Supplementary Fig. S4B and C).
Figure 4. Inhibition of lung cancer cell proliferation and viability by CYT387.
A, Crystal violet stain of A549 cell clonogenic assay after 10 d treatment with DMSO, 2.5 µM MRT67307, CYT387, or Ruxolitinib.
B, Dose-response to MRT67307 or CYT387 treatment at 72 h normalized to DMSO control, mean ± SEM of quadruplicate samples shown. Blue = KRAS mutant/dependent cell lines, Red = KRAS WT/independent cell lines.
C, Cell viability of 12 KRAS dependent (blue) or independent (red) lung cancer lines (purple = KRAS WT/dependent) treated with 5 µM inhibitors for 72 h normalized to DMSO control. Plotted are relative cell line sensitivities for MRT67307/BX795 and AZD6244/CI-1040 (upper panels), or CYT387/MRT67307 and CYT387/AZD6244 (lower panels). * p < 0.05, Pearson correlation.
D, Normalized relative CYT387 sensitivity in the same NSCLC lines compared with 17 other targeted inhibitors at 5 µM. Nearest neighbor analysis by Pearson correlation, * p < 0.05.
E, Phase contrast images (20×) of A549 spheroids co-cultured with HUVECs and treated with DMSO, MRT67307, CYT387, or Ruxolitinib (1 µM) for 18 or 36 h. Mean number and SD of dispersed cells per spheroid from triplicate devices shown.
F, Phase contrast images (20×) of A549 spheroids stimulated with EGF and treated with DMSO or 1 µM CYT387 for 36 h. Mean number and SD of dispersed cells per spheroid from triplicate devices shown.
To extend these findings, we treated a panel of 12 NSCLC cell lines with 18 different kinase inhibitors targeting multiple pathways. Cell lines dependent on KRAS were also dependent on TBK1 with the exception of H1819 cells (19, 20), which instead required IKKε expression for survival (Supplementary Fig. S3A and S4D). As expected, cell line sensitivity to MRT67307 and the related TBK1 inhibitor BX-795 was correlated, similar to the concordance between the MEK inhibitors AZD6244 and CI-1040 (Fig. 4C). KRAS-dependent cell sensitivity to these TBK1 inhibitors was also comparable to MEK inhibitors. Cell line sensitivity to CYT387 significantly correlated with MRT67307, but not AZD6244 (Fig. 4C). Indeed, nearest neighbor analysis showed correlation of CYT387 sensitivity with both BX-795 and MRT67307, despite different off-target effects (Fig. 4D). Similarly, CI-1040 was the nearest neighbor to AZD6244 and the AKT inhibitor MK-2206 was closely related to the PI3K inhibitor GDC0941 (Supplementary Fig. S4E and F), confirming specificity. Together, these findings demonstrated that CYT387 and other TBK1 inhibitors impair KRAS-dependent cell viability in a distinct fashion compared with other targeted inhibitors.
To determine sensitivity of cells in 3D culture, we next treated A549 spheroids with MRT67307, CYT387, or Ruxolitinib. Only CYT387 prevented HUVEC-induced spheroid dispersal at 1 µM concentration (Fig. 4E). Treatment with 1 µM CYT387 also abrogated A549 spheroid proliferation and migration in response to EGF stimulation, in contrast to 1 µM MRT67307 or Ruxolitinib, while combination of MRT67307 and Ruxolitinib inhibited spheroid dispersal (Fig. 4F and Supplementary Fig. S4G). These findings were consistent with the effects of CCL5 and IL-6 neutralization, suggesting a unique potential for CYT387 to disrupt these cytokines in lung cancer cells and suppress KRAS-driven tumorigenicity in vivo.
CYT387 suppresses an autocrine cytokine circuit in KRAS-dependent lung cancer cells
We noted that IKKε over-expression promoted CCL5 and IL-6 production even more potently than TBK1 (Supplementary Fig. S5A). IL-6 mRNA expression also recovered during culture of Tbk1−/− MEFs, coinciding with inducible expression of IKKε (Supplementary Fig. S5B and C). Since STAT3 induces IKKε (25) these findings suggested that disruption of a circuit involving IKKε and JAK signaling was required for effective suppression of IL-6. Indeed, treatment of Tbk1−/− MEFs with Ruxolitinib disrupted this circuit and sustained inhibition of CCL5 and IL-6 expression (Supplementary Fig. S5D). Thus, interruption of CCL5 and IL-6 together required inhibition of TBK1/IKKε and JAK signaling.
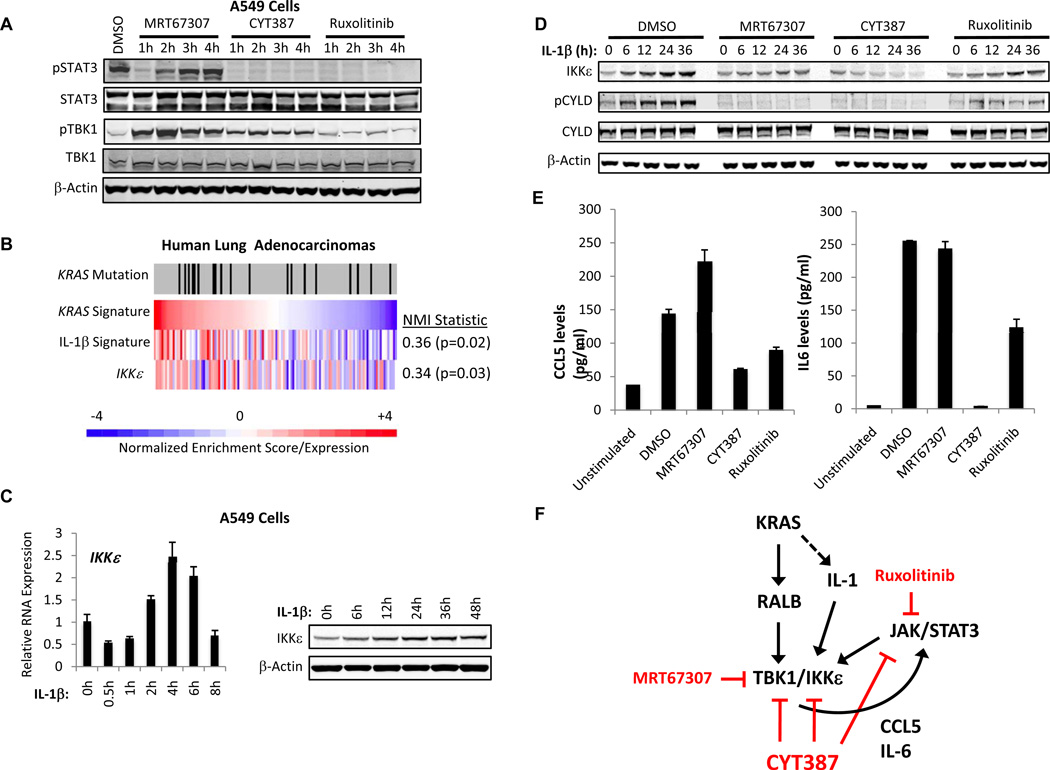
We next assessed the effect of each inhibitor on this autocrine circuit in KRAS-dependent lung cancer cells. Treatment of A549 cells with 5 µM MRT67307 transiently inhibited baseline pSTAT3 levels, but within 2 h led to re-activation of STAT3 consistent with the feedback we observed in Tbk1−/− MEFs (Fig. 5A), as well as paradoxical induction of TBK1 S172 activation. In contrast, treatment with Ruxolitinib inhibited pSTAT3 but had no effect on pTBK1 levels (Fig. 5A). Uniquely, treatment of A549 cells with CYT387 inhibited pSTAT3 and induced pTBK1, to a lesser degree than MRT673037 (Fig. 5A). These findings were consistent with our observations in macrophages and suggested dual targeting of JAK and TBK1 signaling by CYT387 in KRAS-dependent lung cancer cells.
Figure 5. CYT387 treatment disrupts a TBK1/IKKε and JAK-dependent autocrine cytokine circuit.
A, Immunoblot of Y705 pSTAT3, STAT3, S172 pTBK1, TBK1, and β-actin levels in A549 cells treated with 5 µM MRT67307, CYT387, or Ruxolitinib.
B, Heatmap of KRAS/IL-1 signature expression and IKKε gene expression in a panel of human lung adenocarcinomas. KRAS mutation status indicated by tick marks, scale reflects ssGSEA normalized enrichment scores and IKKε gene expression. NMI = normalized mutual information statistic relative to KRAS signature.
C, IKKε mRNA levels and immunoblot of IKKε and β-Actin levels in IL-1β stimulated A549 cells.
D, Immunoblot of IKKε, S418 pCYLD, CYLD or β-actin levels in IL-1β stimulated A549 cells following DMSO, MRT67307, CYT387, or Ruxolitinib (5 µM) treatment.
E, ELISA of CCL5 and IL-6 levels in A549 cells stimulated for 4h with IL-1β following pretreatment for 1 h with DMSO, MRT67307, CYT387, or Ruxolitinib (5 µM). Mean and SD of duplicate samples shown.
F, Model of autocrine cytokine circuit involving TBK/IKKε, CCL5/IL-6, and JAK/STAT activation.
Since IL-1 signaling potentiated TBK1 and CCL5/IL6 activation in A549 cells and promotes KRAS-dependent tumorigenicity (7, 8), we focused on the consequences of inhibitor treatment in this setting. Using single sample gene set enrichment analysis (ssGSEA) of KRAS and IL-1β signatures we confirmed co-activation of oncogenic KRAS and IL-1 signaling in primary human lung adenocarcinomas (p=0.02, normalized mutual information statistic) (19, 35), and IKKε over-expression in these tumors (Fig. 5B). In consonance with this finding, IL-1 also promoted IKKε mRNA and protein expression in A549 cells (Fig. 5C).
Following short-term stimulation with IL-β or IL-6 as a control, CYT387 treatment targeted both pTBK1 and pSTAT3, in contrast to MRT67307 or Ruxolitinib (Supplementary Fig. S5E). CYT387 and MRT67037 treatment but not Ruxolitinib also inhibited basal and IL-1β-induced pCYLD levels, while CYT387 uniquely prevented IL-1β-induced IKKε protein and mRNA expression (Fig. 5D and Supplementary Fig. S5F). MRT67307 treatment failed to suppress IL-1β induced CCL5/IL-6 expression and Ruxolitinib partially reduced these levels of these cytokines, whereas CYT387 treatment strongly inhibited CCL5/IL-6 production (Fig. 5E and Supplementary Fig. S5G). These experiments confirmed that CYT387 targets TBK1/IKKε and JAK signaling in lung cancer cells and disrupts this autocrine cytokine circuit, suppressing CCL5/IL6 and inducible IKKε expression (Fig. 5F).
IKKε re-activation promotes CYT387 resistance
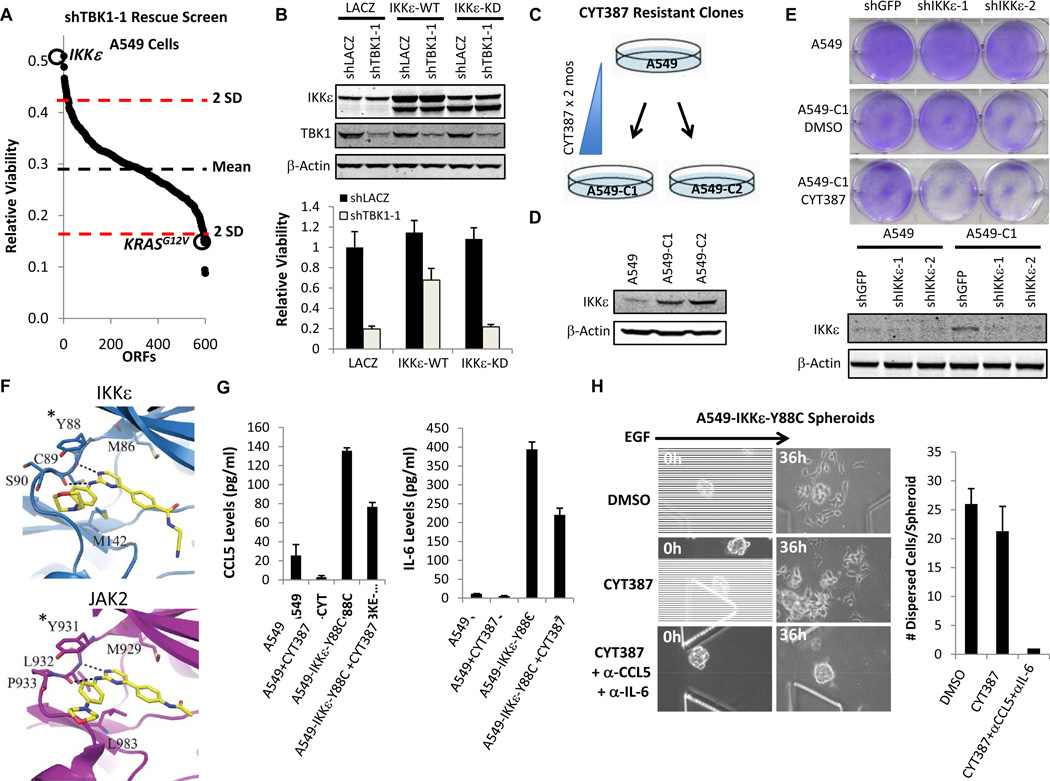
In parallel to these studies, we conducted a kinase open reading frame (ORF) rescue screen to identify genetic modifiers of TBK1 regulated proliferation/survival in A549 cells. We expressed an arrayed library of 600 kinase ORFs in A549 cells (36), then suppressed TBK1 using a TBK1-specific shRNA and measured cell viability. Although we were unable to overexpress TBK1 stably, IKKε was the top-scoring ORF that rescued TBK1 suppression, while oncogenic KRAS enhanced dependency of A549 cells on TBK1 (Fig. 6A, Supplementary Table 1). IKKε-mediated rescue of TBK1 was dependent on its kinase activity, since IKKε WT but not a kinase dead (KD) allele restored A549 cell viability (Fig. 6B).
Figure 6. IKKε-induced CCL5 and IL6 promote CYT387 resistance.
A, Kinase ORF rescue screen of TBK1 suppression in A549 cells. Dotted lines mark 2 SD above and below the mean. Highlighted are IKKε and KRASG12V.
B, Immunoblot of IKKε, TBK1, and β-Actin levels in A549 cells 3 d following TBK1 suppression and IKKε expression (KD = kinase dead). Relative viability after 6 d was normalized to control LACZ expression and suppression. Mean ± SD of 48 replicate samples shown.
C, Schematic of resistant cell line generation over 2 months of CYT387 treatment.
D, Immunoblot IKKε and β-actin levels in parental A549 cells, A549-C1, and A549-C2.
E, Crystal violet stain of A549 or A549-C1 cells stably infected with control shGFP, shIKKε-1 or shIKKε-2 and treated with DMSO or CYT387 for 5 d. Immunoblot shows IKKε and β-actin levels.
F, Model of CYT387 binding to IKKε or JAK2. *homologous IKKε Y88 and JAK2 Y931 residues.
G, ELISA of CCL5 and IL-6 levels in A549 or A549-IKKε-Y88C cells following 24 h treatment with DMSO or CYT387. Mean ± SD of duplicate samples shown.
H, Phase contrast images (20×) of A549-IKKε-Y88C spheroids stimulated with EGF and treated with DMSO or 1 µM CYT387 ± CCL5/IL-6 neutralization for 36 h. Number of cells dispersed per spheroid from triplicate devices shown (Mean ± SD).
We also treated A549 cells with escalating doses of CYT387 for 2 months and identified 2 different clones (A549-C1 and A549-C2) resistant to CYT387 at concentrations as high as 5 µM (Fig. 6C). CYT387 or Ruxolitinib treatment inhibited pSTAT3 in these clones, making it unlikely that drug efflux or mutations in JAK were responsible for resistance (Supplementary Fig. S6A). Instead, each clone expressed substantially increased levels of IKKε relative to parental A549 cells (Fig. 6D). CYT387-resistant A549-C1 cells were resensitized to CYT387 treatment after IKKε suppression (Fig. 6E), suggesting that IKKε reactivation contributes to CYT387 resistance.
To further assess the role of IKKε kinase signaling as a resistance mechanism, we mapped the JAK2 inhibitor resistance alleles Y931C and G935R (37) onto a model of the IKKε kinase domain from the TBK1 structure (38). We noted homologous orientation of JAK2 Y931 and IKKε Y88 (Fig. 6F), suggesting that substitutions involving this residue would block CYT387 binding to IKKε. Indeed, IKKε-Y88C specifically retained kinase activity and rescued CCL5 and IL-6 production following expression and CYT387 treatment in 293T cells (Supplementary Fig. S6B). We then stably expressed IKKε-Y88C, IKKε-WT, IKKε-KD or EGFP in A549 cells and cultured cells in 5 µM CYT387. In contrast to EGFP or IKKε-KD, both IKKε-Y88C and to a lesser extent IKKε-WT accelerated resistance (Supplementary Fig. S6C). Sequencing of IKKε in resistant A549-IKKε-Y88C cells that emerged confirmed dominant expression of the IKKε-Y88C allele (Supplementary Fig. S6D). A549-IKKε-Y88C cells produced high levels of CCL5 and IL-6 insensitive to CYT387 treatment (Fig. 6G). When cultured as spheroids in 3D culture, A549-IKKε-Y88C cells dispersed despite 1 µM CYT387 treatment (Fig. 6H). Addition of CCL5 and IL-6 neutralizing antibodies completely suppressed proliferation, confirming that IKKε-mediated CCL5 and IL-6 production was responsible for CYT387 resistance (Fig. 6H). Together, these findings reveal that inhibition of this autocrine cytokine circuit contributes directly to CYT387 activity in KRAS-dependent lung cancer cells.
Therapeutic effect of CYT387 in murine Kras-driven lung cancers
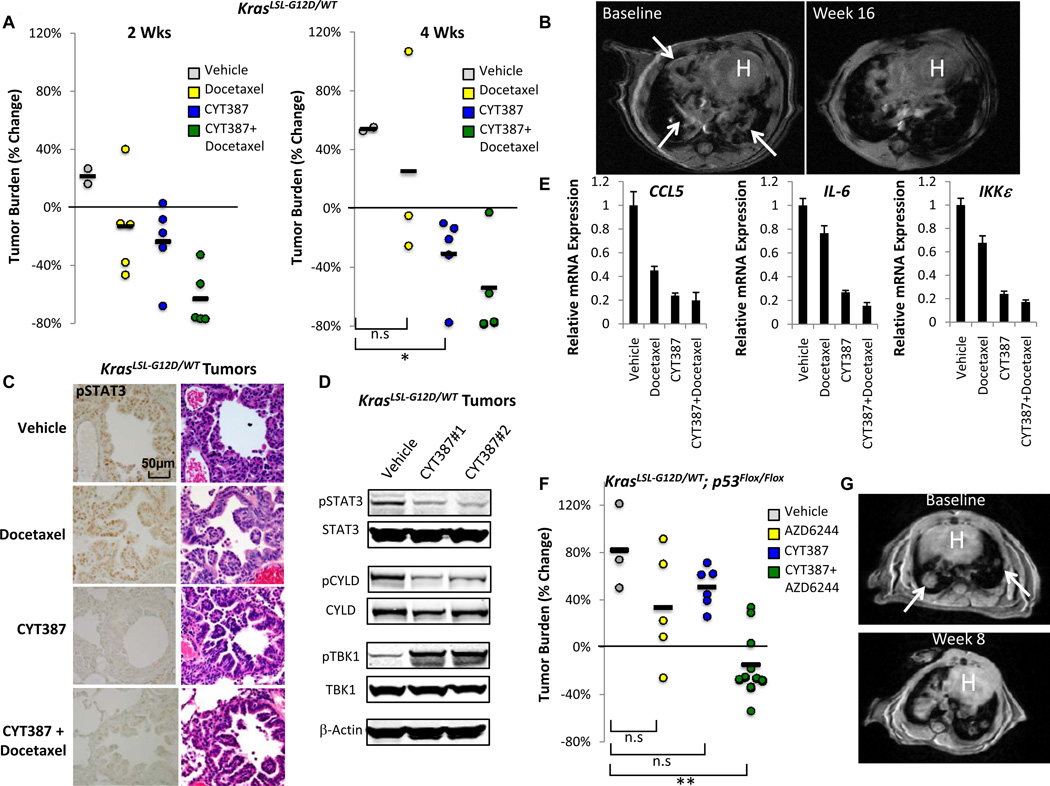
CYT387 inhibits disease progression of JAK-dependent murine myeloproliferative neoplasms and suppresses pSTAT3 in vivo (29). Using the daily dose of CYT387 in these studies as a starting point, we examined CYT387 treatment in a murine model of KrasG12D-driven lung cancer (39). We identified mice with established lung tumors on magnetic resonance imaging (MRI) (40), and treated them with CYT387 100 mg/kg daily by oral gavage, docetaxel 16 mg/kg every other day by intraperitoneal injection, or combination CYT387/docetaxel.
Single agent CYT387 treatment reduced mean tumor volume on MRI as effectively as docetaxel at 2 wks (−23.4% CYT387 vs. −13.0% docetaxel), whereas vehicle treated animals progressed similar to previous studies (40) (Fig. 7A). At 4 wks, CYT387 treatment resulted in statistically significant and durable reduction in mean tumor burden (−30.5%, p<0.01 vs. vehicle, t-test) in contrast to docetaxel (+25.8%, p=0.626 vs. vehicle, t-test) (Fig. 7A). CYT387-induced lung tumor regression was durable even at 16 wks (Fig. 7B). Furthermore, while 2 of the docetaxel treated mice died by 4 wks from toxicity, CYT387-treated mice exhibited no overt signs of toxicity. We observed potent synergy between CYT387 and docetaxel (mean tumor volume reduction −62.8% and −52.8% at 2 and 4 wks respectively for the combination) (Fig. 7A), although this combination resulted in weight loss, which was not observed in mice treated with CYT387 alone (Supplementary Fig. S7A).
Figure 7. Therapeutic efficacy of CYT387 in Kras-driven murine lung cancer.
A, % change in MRI tumor volume of KrasLSL-G12D/WT induced lung cancer 2 and 4 wk following vehicle, docetaxel, CYT387, or CYT387+docetaxel treatment. * p<0.01, t-test.
B, Baseline and 16 wk MRI images following CYT387 treatment. Arrows indicate baseline lung tumor burden. H = heart.
C, Immunohistochemistry for Y705 pSTAT3 levels in tumors from vehicle, docetaxel, CYT387, or CYT387+docetaxel treated mice compared with Hematoxylin-eosin (HE) stain.
D, Immunoblot of Y705 pSTAT3, STAT3, S418 pCYLD, CYLD, S172 pTBK1, TBK1, and β-Actin levels in tumors from vehicle or 2 different CYT387 treated mice.
E, mRNA levels of CCL5, IL6, and IKKε in tumors from docetaxel, CYT387, or CYT387 + docetaxel treated mice, normalized to vehicle control. Mean ± SEM of triplicate samples shown.
F, % change in tumor burden of KrasLSL-G12D/WT; p53Flox/Flox lung cancer treated with vehicle, AZD6244, CYT387, or the combination at 2 wk. Two mice treated with the combination were imaged at 3 wks. ** p<0.001, t-test.
G, Baseline and 8 wk MRI images following CYT387/AZD6244 therapy. Arrows indicate baseline lung tumor burden. H = heart.
To determine whether CYT387 suppressed JAK and TBK1/IKKε signaling in vivo we measured several markers of activity in tumors from treated mice. CYT387 or CYT387/docetaxel treatment specifically inhibited tumor pSTAT3 levels compared with vehicle or docetaxel alone (Fig. 7C and D). TBK1 signaling was active in these tumors as measured by elevated pCYLD and pTBK1 levels, and, in consonance with our findings in vitro, CYT387 treatment blocked CYLD phosphorylation and paradoxically increased pTBK1 (Figure 7D). As a consequence, levels of CCL5, IL-6, and IKKε were all preferentially reduced in CYT387 and CYT387/docetaxel treated animals (Fig. 7E). Thus, response to CYT387 therapy correlated with effective disruption of this cytokine circuit in vivo.
MAPK pathway inhibition is a central focus of current therapeutic strategies targeting KRAS-driven cancers. Using a phospho-MAPK antibody array, we confirmed that CYT387 treatment in vitro failed to suppress MAPK pathway activation in KRAS-dependent A549 cells, and instead increased pERK1/2 levels compared with Ruxolitinib or with AZD6244, which inhibited MEK-ERK signaling as expected (Supplementary Fig. S7B). Treatment with either MRT67307 or CYT387 rapidly induced feedback pERK activation in A549 cells (Supplementary Fig. S7C), also reported following TBK1 suppression in this cell line (33). Inhibition of this feedback by co-treatment with AZD6244 and MRT67307 blocked cytokine expression, and the combination of AZD6244 and CYT387 completely ablated basal IL-6 production (Supplementary Fig. S7D). Treatment of KRAS dependent cell lines with AZD6244 and CYT387 together prevented pSTAT3 and pERK1/2 activation, indicating that combined TBK1/IKKε, JAK, and MEK inhibition suppressed these multiple KRAS-driven pathways (Supplementary Fig. S7E), resulting in significantly greater cell death compared to either inhibitor alone (Supplementary Fig. S7F). Building on these experiments, we next explored this combination in murine KrasLSL-G12D/WT-p53flox/flox (KP) lung cancer (41), an aggressive model with high levels of pERK1/2 (40). In contrast to treatment with either agent alone, combined CYT387/AZD6244 therapy of established KP tumors resulted in statistically significant tumor regression (−14.4%, p<0.001 vs. vehicle, t-test) (Fig. 7F). Furthermore, we observed no overt drug-related toxicity and minimal disease progression in responding mice even at 8 weeks (Fig. 7G and Supplementary Fig. S7G). Thus, suppression of multiple RAS effector pathways by combination CYT387/AZD6244 treatment not only impairs viability of KRAS-dependent NSCLC cells in vitro, but also inhibits tumor growth in the treatment refractory KP lung cancer model.
DISCUSSION
In addition to their key roles in innate immunity, TBK1 and IKKε have also been implicated in malignant transformation (42), basal autophagy (22) and obesity (43). These observations suggest that TBK1 and IKKε likely have multiple substrates whose phosphorylation regulates specific functions in a context- and cell type-dependent manner.
Oncogenic KRAS co-opts TBK1/IKKε signaling by activating RALB (18) and inducing IKKε expression (23). Yet the mechanism by which TBK1 and IKKε regulate transformation and cancer cell survival remains incompletely characterized. Here we found that CCL5 and IL-6, key TBK1/IKKε associated cytokines during innate immunity, are also aberrantly activated in KRAS-driven lung cancers. These cytokines, in turn, act in an autocrine manner to activate JAK/STAT signaling, which amplifies IL-6 production, promotes IKKε expression, and sustains growth factor activated lung cancer cell proliferation and migration. We identified CYT387, a clinical stage JAK inhibitor, as a potent TBK1/IKKε inhibitor that disrupts this autocrine loop and causes regression of established murine Kras-driven lung tumors in vivo. These observations elucidate a novel KRAS effector pathway emanating from RAL-GEF through RALB and TBK1 that activates cytokine production in an inappropriate context to promote tumor maintenance. Since genetic and pharmacologic perturbation of these kinases extinguishes this cytokine signaling network and inhibits KRAS-driven tumorigenicity, targeting this effector pathway represents a promising new strategy for these treatment refractory cancers.
Contributions of inflammatory signaling to KRAS-induced tumorigenesis
Oncogenic KRAS constitutively activates multiple effectors, including MAPK, PI3K and RAL-GEF. Although most cancers that harbor oncogenic KRAS mutations also depend on KRAS signaling, KRAS mutation and dependency are not strictly correlated (19, 31), indicating that RAS signaling is active and required other contexts as well. The finding that TBK1 links RALB to the generation of specific protumorigenic cytokines provides mechanistic insights into the factors that drive KRAS induced cell transformation. CCL5-CCR5 signaling promotes chemotaxis and enhances metastasis of RAS-driven breast cancer cells (44). Multiple studies have established that IL-6 and STAT3 induced survival signaling are required for RAS-mediated tumorigenesis (10–13). TBK1/IKKε regulated CCL5 and IL-6 influence the local inflammatory microenvironment and may also support tumorigenesis in a non-cell autonomous fashion (45). Activation of this pathway is conversely influenced by factors in the tumor microenvironment and thus may occur in other contexts, although our observations demonstrate that TBK1 is activated and required by lung cancer cells that depend on KRAS. KRAS independent cancers that exhibit little activation of TBK1 and cytokine signaling may maintain tumorigenesis via alternative means.
These studies identified an autocrine cytokine circuit involving TBK1/IKKε and JAK activity that sustained CCL5 and IL-6 production in cancer cells, similar to IFN-β signaling (26). Like IFN-β, both CCL5 and IL-6 activate JAK/STAT signaling, inducing IKKε expression (25) and triggering further CCL5 and IL-6 production (Fig. 5F). TBK1 inactivation and JAK inhibition together were required to ablate CCL5 and IL-6 production in MEFs, and inhibition of TBK1/IKKε and JAK by CYT387 was necessary to inhibit induction of IKKε, CCL5 and IL-6 in KRAS-dependent lung cancer cells. IKKε over-expression rescued TBK1 suppression, CYT387 resistant clones overexpressed IKKε, and an inhibitor resistant IKKε allele conferred resistance to CYT387, suggesting that these closely related kinases are key targets of CYT387. Although CYT387 inhibits IKKε kinase activity, IKKε may have a scaffolding function or require maximal ATP-site occupancy for full inhibition, which is more difficult in the presence of excess kinase and has been observed for other enzymatic drug targets such as DHFR (46). The requirement of CCL5 and IL-6 for IKKε-driven resistance confirms the involvement of this autocrine signaling loop in KRAS-dependent lung cancer proliferation.
TBK1/IKKε also activate AKT (20, 21) and regulate NF-κB signaling components such as CYLD (19, 30). Rescue of Tbk1−/− MEF clonogenic proliferation by CCL5 or conditioned media was incomplete, suggesting that cell intrinsic defects in AKT, NF-κB signaling or autophagy (22) also contribute to TBK1-regulated cell survival. Indeed, MRT67307 treatment impaired cell viability at higher concentrations, suggesting that TBK1 likely counterbalances oncogenic stress by additional means. Given the emerging link between autophagy and RAS-driven tumorigenesis (47), it will be interesting to elucidate the interface between TBK signaling and this particular stress-response pathway in KRAS-dependent cancer cells.
Implications for therapy of RAS-driven cancer
CYT387 treatment of murine Kras-driven lung cancers was well tolerated and suppression of tumor growth was more durable than standard docetaxel chemotherapy. Combined CYT387 and AZD6244 treatment exhibited significant therapeutic activity in KrasLSL-G12D/WT-p53Flox/Flox-induced murine lung cancers, a treatment-refractory model. Since CYT387 and MEK inhibitors are in advanced stages of clinical development, these observations provide a rationale for testing this combination in human KRAS-driven cancers.
Although kinase inhibitor selectivity is desirable, the complexity of solid tumors and cancer cell signal transduction necessitates concomitant inhibition of several signaling pathways (48). For example, multi-targeted RTK inhibitors are clinically effective against a variety of tumors. Clinical trials of CYT387 in myelofibrosis have also demonstrated safety and efficacy unique among JAK inhibitors in improving transfusion dependence (49). Thus, multi-targeted kinase inhibition need not correlate with enhanced toxicity. Finally, since RALB-TBK1 signaling is distinct from MAPK and PI3K, the observation that CYT387 treatment synergizes with MEK inhibition suggests that further combination strategies may be effective in treating KRAS-driven cancers and overcoming resistance.
METHODS
Detailed protocols for all sections are described in Supplementary Methods
Cell Culture
Cells were cultured at 37°C in a humidified incubator with 5% CO2. NSCLC cell lines were obtained from ATCC or the DFCI-84 collection, and cell identity by KRAS mutation status was previously validated by sequencing (19). Murine RAW 264.7 gamma NO(−) cells and HUVECs were from ATCC. Tbk1−/− and WT MEFs were kindly provided by K. Fitzgerald. Tumor spheroids were generated from A549 or NCI-H1437 cells using low attachment plates and embedded in a 3D microfluidic culture system (34).
Gene transduction
Cells nucleofection/transfection (38) or introduction of shRNAs/ORFs by lentiviral transduction was performed as described (19, 36). shRNA sequences are listed in Supplementary Table 2.
Immunoblotting, Antibodies, and ELISA
Immunoblotting was performed as described (19). Cytokines, ELISA kits and cytokine antibody arrays were purchased from R&D Systems.
In-vitro enzyme kinetic assays
Recombinant enzymes were incubated with ATP and IKKε tide (ADDDYDSLDWDAKKK) as a specific TBK1/IKKε peptide substrate as described (30).
Quantitative Real-Time PCR
Real-time PCR was performed using LightCycler® 480 SYBR Green I Master (Roche). The sequences of the primers used for RT-PCR are listed in Supplementary Table 3.
Animal Studies
Mouse experiments were conducted in agreement with a protocol approved by the Institutional Animal Care and Use Committee (IACUC). Lung tumors in KrasLSL-G12D/WT or KrasLSL-G12D/WT; p53Flox/Flox mice were induced by inhalation of adenoviral Cre recombinase and monitored by serial MRI scanning as described (40).
Immunohistochemical Analysis
IHC staining was performed on formalin fixed, paraffin embedded tissues from treated mice sectioned at 5 µm thickness.
Supplementary Material
SIGNIFICANCE.
Oncogenic KRAS engages cytokine signaling, in addition to MAPK and PI3K activation, to promote tumorigenesis. CYT387, originally described as a selective JAK inhibitor, is also a potent TBK/IKKε inhibitor that uniquely disrupts a cytokine circuit involving CCL5 and IL-6 and STAT3. The efficacy of CYT387-based treatment in murine Kras-driven lung cancer models uncovers a novel therapeutic approach for these refractory tumors with immediate translational implications.
Acknowledgements
We thank C. Yu, J. Boehm, A. Stern, T. Sim, Q. Liu, and N. Gray for helpful discussions, and R. Sharifi Sedeh for assistance with 3D culture analysis.
Grant support: This work is supported by U.S. NIH K08 CA138918-01A1 (DAB), P01 CA154303 and R01 CA130988 (WCH), and R01 CA122794, R01 CA166480, R01 CA163896, R01CA140594, and U01CA141576 (KW). DAB is supported by V Scholar, Gloria Spivak Faculty, GTM Fund for Lung Cancer Research, and Friends of the Dana-Farber Cancer Institute Awards. ZZ and KW are supported by Uniting Against Lung Cancer Grants.
Footnotes
Disclosure of Potential Conflicts of Interest: JAE, MJE and WCH are consultants for Novartis. DAB is a consultant for N-of-One.
References
- 1.Downward J. Targeting RAS signalling pathways in cancer therapy. Nature reviews Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 2.Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes & development. 2002;16:2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayo MW, Wang CY, Cogswell PC, Rogers-Graham KS, Lowe SW, Der CJ, et al. Requirement of NF-kappaB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 6.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, et al. The GATA2 Transcriptional Network Is Requisite for RAS Oncogene-Driven Non-Small Cell Lung Cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 8.Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, et al. KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer cell. 2012;21:105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 10.Ancrile B, Lim KH, Counter CM. Oncogenic Ras-induced secretion of IL6 is required for tumorigenesis. Genes & development. 2007;21:1714–1719. doi: 10.1101/gad.1549407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer cell. 2011;19:456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Corcoran RB, Contino G, Deshpande V, Tzatsos A, Conrad C, Benes CH, et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer research. 2011;71:5020–5029. doi: 10.1158/0008-5472.CAN-11-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda A, Wang SC, Morris JPt, Folias AE, Liou A, Kim GE, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 15.Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer cell. 2004;6:447–458. doi: 10.1016/j.ccr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 16.Wislez M, Fujimoto N, Izzo JG, Hanna AE, Cody DD, Langley RR, et al. High expression of ligands for chemokine receptor CXCR2 in alveolar epithelial neoplasia induced by oncogenic kras. Cancer research. 2006;66:4198–4207. doi: 10.1158/0008-5472.CAN-05-3842. [DOI] [PubMed] [Google Scholar]
- 17.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nature reviews Cancer. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 18.Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 19.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ou YH, Torres M, Ram R, Formstecher E, Roland C, Cheng T, et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Molecular cell. 2011;41:458–470. doi: 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie X, Zhang D, Zhao B, Lu MK, You M, Condorelli G, et al. IkappaB kinase epsilon and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6474–6479. doi: 10.1073/pnas.1016132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AC, Scholefield CL, Kemp AJ, Newman M, McIver EG, Kamal A, et al. TBK1 Kinase Addiction in Lung Cancer Cells Is Mediated via Autophagy of Tax1bp1/Ndp52 and Non-Canonical NF-kappaB Signalling. PloS one. 2012;7:e50672. doi: 10.1371/journal.pone.0050672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajurkar M, De Jesus-Monge WE, Driscoll DR, Appleman VA, Huang H, Cotton JL, et al. The activity of Gli transcription factors is essential for Kras-induced pancreatic tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1038–E1047. doi: 10.1073/pnas.1114168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 25.Guo J, Kim D, Gao J, Kurtyka C, Chen H, Yu C, et al. IKBKE is induced by STAT3 and tobacco carcinogen and determines chemosensitivity in non-small cell lung cancer. Oncogene. 2013;32:151–159. doi: 10.1038/onc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Tenoever BR, Sharma S, Zou W, Sun Q, Grandvaux N, Julkunen I, et al. Activation of TBK1 and IKKvarepsilon kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. Journal of virology. 2004;78:10636–10649. doi: 10.1128/JVI.78.19.10636-10649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu GY, He G, Li CY, Tang M, Grivennikov S, Tsai WT, et al. Hepatic expression of HCV RNA-dependent RNA polymerase triggers innate immune signaling and cytokine production. Molecular cell. 2012;48:313–321. doi: 10.1016/j.molcel.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark K, Peggie M, Plater L, Sorcek RJ, Young ER, Madwed JB, et al. Novel cross-talk within the IKK family controls innate immunity. Biochem J. 2011;434:93–104. doi: 10.1042/BJ20101701. [DOI] [PubMed] [Google Scholar]
- 29.Tyner JW, Bumm TG, Deininger J, Wood L, Aichberger KJ, Loriaux MM, et al. CYT387, a novel JAK2 inhibitor, induces hematologic responses and normalizes inflammatory cytokines in murine myeloproliferative neoplasms. Blood. 2010;115:5232–5240. doi: 10.1182/blood-2009-05-223727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutti JE, Shen RR, Abbott DW, Zhou AY, Sprott KM, Asara JM, et al. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Molecular cell. 2009;34:461–472. doi: 10.1016/j.molcel.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival. Cancer cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JY, Welsh EA, Oguz U, Fang B, Bai Y, Kinose F, et al. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12414–12419. doi: 10.1073/pnas.1220674110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aref AR, Huang RY, Yu W, Chua KN, Sun W, Tu TY, et al. Screening therapeutic EMT blocking agents in a three-dimensional microenvironment. Integrative biology : quantitative biosciences from nano to macro. 2013;5:381–389. doi: 10.1039/c2ib20209c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jura J, Wegrzyn P, Korostynski M, Guzik K, Oczko-Wojciechowska M, Jarzab M, et al. Identification of interleukin-1 and interleukin-6-responsive genes in human monocyte-derived macrophages using microarrays. Biochimica et biophysica acta. 2008;1779:383–389. doi: 10.1016/j.bbagrm.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weigert O, Lane AA, Bird L, Kopp N, Chapuy B, van Bodegom D, et al. Genetic resistance to JAK2 enzymatic inhibitors is overcome by HSP90 inhibition. The Journal of experimental medicine. 2012;209:259–273. doi: 10.1084/jem.20111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tu D, Zhu Z, Zhou AY, Yun CH, Lee KE, Toms AV, et al. Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell reports. 2013;3:747–758. doi: 10.1016/j.celrep.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 40.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–617. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer research. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 42.Shen RR, Hahn WC. Emerging roles for the non-canonical IKKs in cancer. Oncogene. 2011;30:631–641. doi: 10.1038/onc.2010.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reilly SM, Chiang SH, Decker SJ, Chang L, Uhm M, Larsen MJ, et al. An inhibitor of the protein kinases TBK1 and IKK-varepsilon improves obesity-related metabolic dysfunctions in mice. Nature medicine. 2013;19:313–321. doi: 10.1038/nm.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 45.Czabanka M, Korherr C, Brinkmann U, Vajkoczy P. Influence of TBK-1 on tumor angiogenesis and microvascular inflammation. Frontiers in bioscience : a journal and virtual library. 2008;13:7243–7249. doi: 10.2741/3225. [DOI] [PubMed] [Google Scholar]
- 46.Carman MD, Schornagel JH, Rivest RS, Srimatkandada S, Portlock CS, Duffy T, et al. Resistance to methotrexate due to gene amplification in a patient with acute leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1984;2:16–20. doi: 10.1200/JCO.1984.2.1.16. [DOI] [PubMed] [Google Scholar]
- 47.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes & development. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nature reviews Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardanani A, Laborde RR, Lasho TL, Finke C, Begna K, Al-Kali A, et al. Safety and efficacy of CYT387, a JAK1 and JAK2 inhibitor, in myelofibrosis. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2013;27:1322–1327. doi: 10.1038/leu.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.