SUMMARY
Molecular characterization studies of a diverse collection of avian influenza viruses (AIVs) have demonstrated that AIVs’ greatest genetic variability lies in the HA, NA, and NS genes. The objective here was to quantify the association between geographical locations, periods of time, and host species and pairwise nucleotide variation in the HA, NA, and NS genes of 70 isolates of H5N1 highly pathogenic avian influenza virus (HPAIV) collected from October 2005 to December 2007 from birds in Romania. A mixed-binomial Bayesian regression model was used to quantify the probability of nucleotide variation between isolates and its association with space, time, and host species. As expected for the three target genes, a higher probability of nucleotide differences (odds ratios [ORs] > 1) was found between viruses sampled from places at greater geographical distances from each other, viruses sampled over greater periods of time, and viruses derived from different species. The modeling approach in the present study maybe useful in further understanding the molecular epidemiology of H5N1 HPAI virus in bird populations. The methodology presented here will be useful in predicting the most likely genetic distance for any of the three gene segments of viruses that have not yet been isolated or sequenced based on space, time, and host species during the course of an epidemic.
Keywords: Romania, H5N1 highly pathogenic avian influenza virus, space, time, birds, molecular epidemiology
The greatest genetic variability of avian influenza virus’s (AIV’s) eight-segmented RNA genome is found in the HA, NA, and NS genes (16). The HA and NA genes encode the two major AIV surface proteins. The HA glycoprotein mediates virus attachment to host-cell surface receptors. It is the major antigen against which host-neutralizing antibodies are developed (5,25,27). The NA gene encodes another surface protein, which is another antigenic determinant and has enzymatic activity that enables virus progeny release from infected cells, thus facilitating spread (1,25). The NS gene encodes the nonstructural protein that is associated with differential virus replication kinetics, pathogenicity, and host range restriction and adaptation (18,19,32).
The emergence of the H5N1 HPAIV ancestral strain A/goose/Guangdong/1/96 occurred in commercial domesticated geese in Guandong province in China in 1996 (36). Since then, the ancestral strain has undergone numerous genetic changes leading to the evolution of different lineages (or clades) (17). To date, 10 clades of H5N1 HPAIV have been identified based on sequence comparisons of hemagglutinin genes of 859 H5N1 sequences (35). H5N1 HPAIVs are now spread to a number of countries in Asia, Europe, and Africa. The H5N1 subclade 2.2 emerged between 2004 and 2005, and caused outbreaks in wild birds and poultry in northwestern China before spreading out of Asia and westwards into Russia and Europe in 2005 (4,17). Romania was one of the first countries to report H5N1 HPAIV subclade 2.2 infection outside of Asia. The first outbreak of H5N1 HPAIV was detected near Golovita Lake in Tulcea County, in southeastern Romania in early October 2005, where the virus was isolated from domesticated chickens and wild birds (34). A total of 69 outbreaks were reported in Romania during 2005 (34). From January 2006 to December 2007, approximately 110 H5N1 HPAIV outbreaks were detected and reported across Romania in both domesticated poultry premises and wild birds (3,34). The outbreaks were predominantly detected in the southeastern part of the country, including the Black Sea coast and river deltas. Phylogenetic analysis of the first H5N1 HPAIV isolates in 2005 indicated that the virus is directly related to the Chinese and Russian isolates (21). Both movement of poultry and wild bird migration appear to play a role in the spread of H5N1 HPAIVs over long distances and into Europe (16,31). Furthermore, it was found that the Romanian H5N1 HPAIV outbreaks were a result of multiple introductions, in which the initial introduction may have been caused by wild birds in the Danube Delta region (9).
Although the genetic and antigenic evolution and geographic spread of the H5N1 HPAIVs was well documented after the initiation of systemic surveillance in 2000 (13), there is little understanding on how genetic variation of H5N1 HPAIV might be influenced by host and other space, time, and host species. The objective of the study was to quantify the association between pairwise nucleotide variation within the HA, NA, and NS genes with space, time, and species of the host population with the use of 70 isolates of H5N1 HPAIV collected from October 2005 to December 2007 from birds in Romania. Results will contribute to knowledge on the molecular epidemiology of the AIV and its associations with host and environmental factors.
MATERIALS AND METHODS
Data
In the present study, 70 H5N1 HPAIVs were collected in Romania as part of the AI surveillance activities in poultry and wild birds conducted by the European Union (EU) member states since 2005 (3). Clinical samples comprised cloacal and/or oral swabs or tissue samples collected in brain–heart infusion with antibiotics media. Viral RNA was isolated from swab eluent or egg-grown isolates with the use of RNA extraction methods as described previously (3). Full genome sequencing was carried out as part of a Wellcome Trust–funded influenza sequencing pipeline initiative. RNA was reverse transcribed and each of the eight gene segments PCR amplified with the use of universal primers (37). Amplicons were then full-genome sequenced with the use of 454 technology and assembled with the use of runMapping software as described elsewhere (12). Sequences and their associated data (date, location, and species of isolation) were uploaded to the secure servers of a Web-based system, referred to as the Disease BioPortal (http://fmdbioportal.ucdavis.edu/). The phylogenetic trees of the HA, NA, and NS gene segments, locations, and reporting dates of the sequenced isolates were visualized with the use of the Disease BioPortal time-space-genomic visualizer. A description of the technical attributes and capabilities of the Disease BioPortal along with an illustration of its functionality are available elsewhere (23,24). A video demonstration of the findings using the Disease BioPortal and including the simultaneous visualization of the geographical, temporal, and genetic relation of isolates is publicly accessible at (http://cadms.ucdavis.edu/dl/bp_vids/Bioportal_Romania_H5N1.mov.zip). Furthermore, three phylogenetic trees were calculated for each gene region to assess the degree of relatedness between the virus isolates. The phylogenetic trees were constructed with Molecular Evolutionary Genetic Analysis software version 5.05 (28) with the use of neighbor-joining tree analysis. A Tamura-Nei γ model was used with 1000 bootstrap replications to assign confidence level to the tree topology.
The pairwise association of the space, time, and host species with nucleotide changes was assessed for H5N1 HPAIVs from any two infected birds (i and j) from 2005 to 2007 in Romania. Predictors were
- KMi,j = pairwise spatial distance (km), which was measured by using the Haversine formula, which gives the great-circle distances between two points on a sphere of radius R, based on the longitude (long) and latitude (lat) of the points, so that:
where R = 6367 km (i.e., the radius of the Earth), and latitude (lat) and longitude (long) were measured in radians. Ti,j = pairwise absolute difference in time (days) between the sampling dates, which was estimated as = | Ti – Tj |.
Spi,j = pairwise difference in the species of isolation, which as encoded as 0 if the sample was isolated from the same bird species, and as 1 if otherwise. Table 1 summarizes the frequency of species of birds tested positive by virus isolation for H5N1 HPAIV during multiple outbreaks between 2005 and 2007 in Romania.
Table 1.
Frequency of species of birds tested positive by virus isolation for H5N1 HPAIV during multiple outbreaks between 2005 and 2007 in Romania.
| Common name | Scientific name | Bird type | Frequency |
|---|---|---|---|
| Chicken | Gallus gallus domesticus | Domestic | 51 |
| Duck | Anas platyrhynchos | Domestic | 5 |
| Guinea hen | Numida meleagris | Wild | 1 |
| Moorhen | Gallinula tenebrosa | Wild | 1 |
| Pigeon | Columba livia | Wild | 1 |
| Muted swan | Cygnus olor | Wild | 7 |
| Turkey | Meleagris gallopavo | Domestic | 3 |
| Canada goose | Branta canadensis | Wild | 1 |
Model formulation
The three gene segments of H5N1 HPAIV were initially aligned with the use of the Clustal X Multiple Sequence Alignment Program, version 1.81 (29). The final alignments were made manually and number of different nucleotides (dn) matrix was calculated with the use of BioEdit Program, version 7.0.9 (8). A mixed Bayesian regression model aimed to quantify the association between genomic differences of virus isolates obtained from diseased animal populations and the variation in space, time, and host species of the reported outbreaks has been described elsewhere (22). Briefly, the outcome of interest was, alternatively, the dni,j in the HA, NA, and NS genes, which were calculated for each pair of isolates (i,j) from all possible pairs of the 70 isolates (N = 2415). In the Bayesian regression model, dni,j was assumed to follow a binomial (n,p) process in which n was the total number of aligned nucleotides compared in each pair of isolates for each gene segment (n = 1754 for HA gene, n = 1371 for the NA gene, and n = 846 for NS gene), and p was the probability that a nucleotide differs between the two HA, NA, NS gene sequences, which, when presented as a percentage, is referred to as the genetic distance. Each predictor (time, distance, species) was incorporated into the model as an independent fixed (f) effect, whereas a nonstructured random effect (U) was included in the formulation to account for lack of independence in the observations due to variables other than the three fixed factors assessed here. Therefore, the model was formally expressed as dn ~ Binomial(n,p), ln[p/(1 – p)] = α+βf+ U, where α denotes the model intercept and β denotes the regression coefficients for each predictor.
Because no prior information exists about the associations between the predictors and genetic distance for the three gene segments, noninformative prior distributions of the form N(0,0.01) and N[0, δ ~ gamma (0.05, 0.005)] were used to model prior knowledge on the value of the regression coefficients for fixed and random effects, respectively. The model was run with the use of the WinBUGS (Bayesian inference by using Gibbs sampling) software, version 1.43 (14), with 20,000 iterations after burning out the first 1000 iterations. Values of KMi,j and Ti,j were standardized by subtracting their value to the variable mean and dividing the results by the variable standard deviation. Variable transformations, including square root, power, exponential and log, were tested to assess whether they improved the model fitness; all possible two-way interactions were also assessed. For each gene segment, the formulation that included variables with significant probability interval for the posterior distribution of its regression coefficient β, and with the smallest deviance information criterion, was considered the model that best fitted the data. The odds ratios (ORs) of each predictor, which were estimated as the exponential of the posterior distribution of the regression coefficients of each significant variable, was computed to quantify the strength of association between factors f and p.
Model validation
For model validation, a split-sample cross-validation approach was used to evaluate the reliability of the future prediction of genetic distance of the model for each of the gene segments. The data set for each possible pair of the 70 isolates H5N1 HPAIV (N = 2415) was randomly partitioned into two groups with the use of a binary random number generator; 60% of the data set (n = 1433) was randomly assigned into a learning subset, whereas the remaining 40% of the data set (n = 982) was randomly assigned into a validation subset. Models were formulated with the use of the learning sample subset and the formulation that best fit the data was used to estimate the predicted genetic distance (PD) in the validation subset. The accuracy of the model was evaluated by computing the Spearman correlation coefficient (R), between the values of PD and the corresponding observed genetic distance (OD) in the validation subset. Furthermore, the sample squared multiple correlation (R2) was estimated by squaring the correlation coefficient (R) between the values of PD and the corresponding OD for each gene segment in the training and validation subset. The quantity R2 of the training subset was subtracted by the R2 of the validation subset to estimate the absolute value of the shrinkage on cross-validation (R*) to assess model reliability in predicting genetic distances for each gene segment. The fitted model was said to be unreliable if R* ≥ 0.9. In contrast, if R* was ≤0.1, then the model was assumed to be reliable. The Grubbs test was used to identify outliers in the correlation of the validation subset, in which an outlier was interpreted to indicate failure to predict genetic distances (6). In addition, a paired t-test was used to assess whether there was a tendency of the model to underestimate or overestimate the genetic distance systematically, by testing the mean difference between the OD and PD in the validation subset. A p value of ≥0.05 was interpreted to indicate no over- or underestimation of the genetic distance for each gene segment.
RESULTS
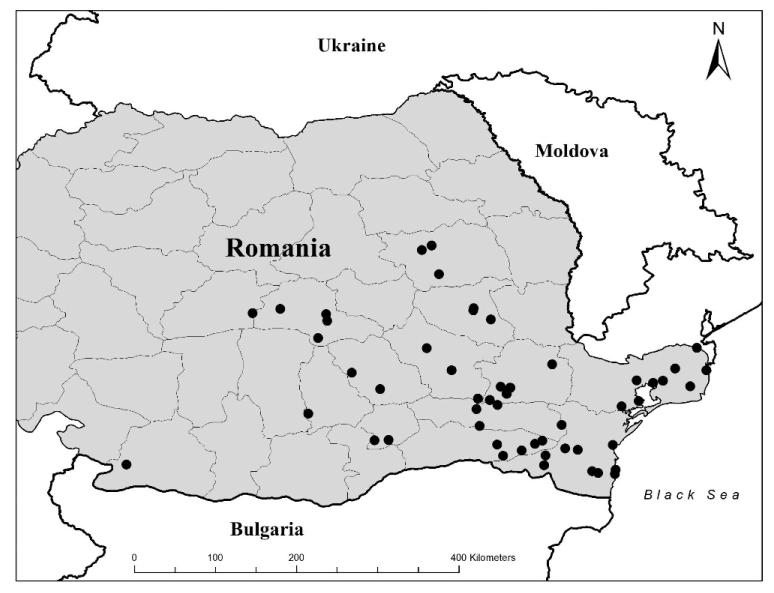
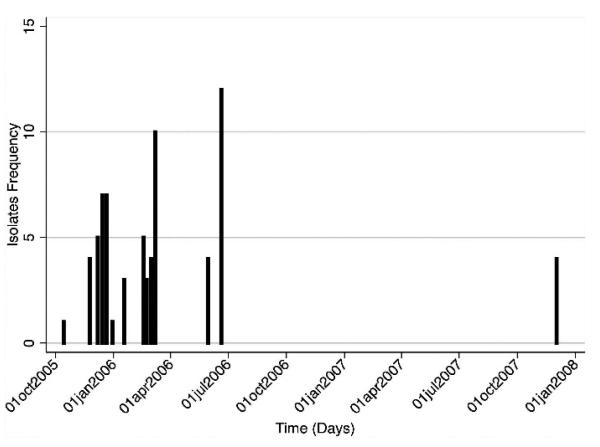
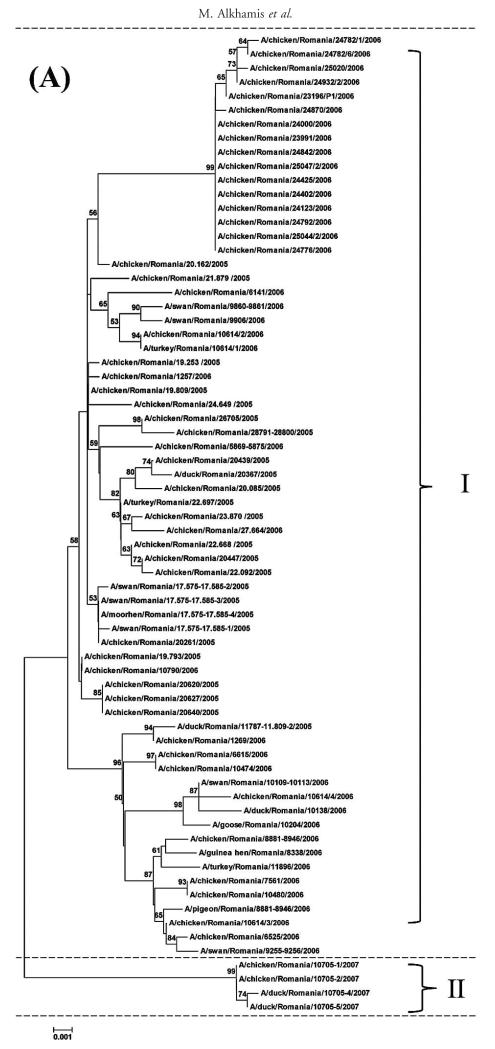
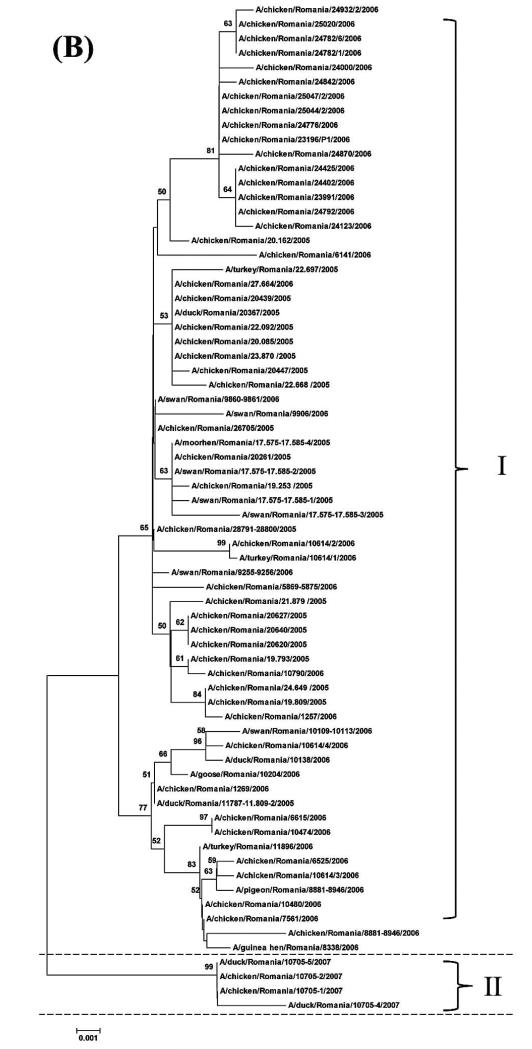
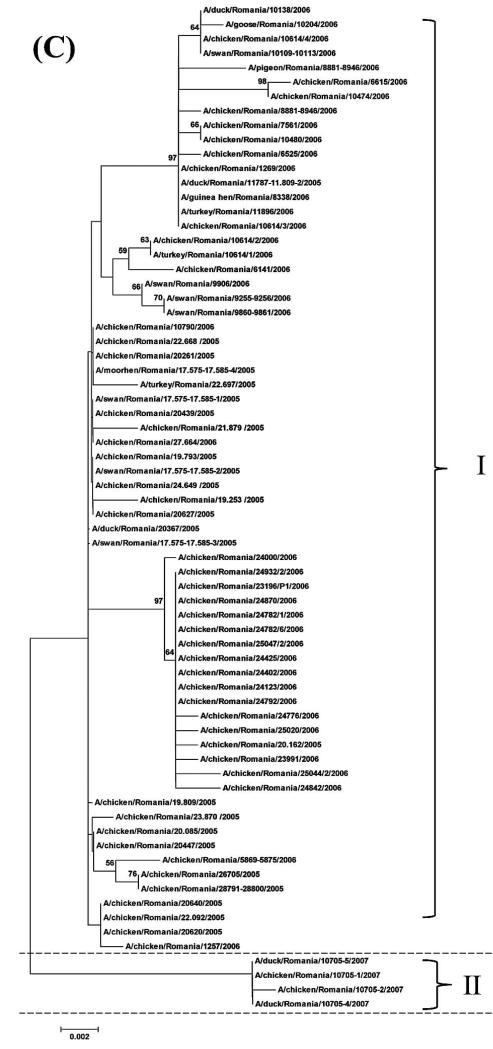
For all the 70 H5N1 HPAIVs, the median numbers of nucleotides that differed among the 2415 pairwise comparisons for the HA, NA, and NS gene segments were 17, 8, and 5, respectively. The minimum and maximum numbers of differing nucleotides were 0 and 43 for the HA gene, 0 and 30 for the NA gene, and 0 and 21 for the NS gene, respectively. The median distance for the 2415 pairwise comparisons, i.e., distance between each pair of isolates, was 137.3 km (1 standardized unit = 89.4 km) with minimum and maximum distances of 0 and 552.4 km, respectively (Fig. 1). The median time was 84 days (1 standardized unit = 188.3 days) with a minimum and maximum time of 0 and 779 days, respectively (Fig. 2). Sequencing and phylogenetic analysis of 70 H5N1 HPAIV in Romania suggest the presence of at least two different groups of viruses. The first group comprised 66 closely related viruses isolated from the 2005–2006 outbreaks, and the second group comprised four closely related viruses isolated from the 2007 outbreak (Fig. 3).
Fig. 1.
Geographical distribution of 70 H5N1 HPAIV isolates sampled from birds during multiple outbreaks between 2005 and 2007 in Romania.
Fig. 2.
Temporal distribution of H5N1 HPAIV isolates sampled from birds during multiple outbreaks between 2005 and 2007 in Romania.
Fig. 3.
(A) Phylogenetic relationship of HA gene of 70 H5N1 HPAIV isolated from sampled birds during multiple outbreaks between 2005 and 2007 in Romania. The phylogenetic trees were generated by neighbor-joining analysis with Tamura-Nei γ model, with the use of MEGA 5.05. Numbers above key nodes indicate the percentage of bootstrap values of 1000 replicates. Group I indicates viruses isolated in 2005–2006, and Group II indicates viruses isolated in 2007. (B) Phylogenetic relationship of NA gene of 70 HPAIV isolated from sampled birds during multiple outbreaks between 2005 and 2007 in Romania. The phylogenetic trees were generated by neighbor-joining analysis with Tamura-Nei γ model, with the use of MEGA 5.05. Numbers above key nodes indicate the percentage of bootstrap values of 1000 replicates. Group I indicates viruses isolated in 2005–2006, and Group II indicates viruses isolated in 2007. (C) Phylogenetic relationship of NS genes of 70 H5N1 HPAIV isolated from sampledbirds during multiple outbreaks between 2005 and 2007 in Romania. The phylogenetic trees were generated by neighbor-joining analysis with Tamura-Nei γ model, with the use of MEGA 5.05. Numbers above key nodes indicate the percentage of bootstrap values of 1000 replicates. Group I indicates viruses isolated in 2005–2006, and Group II indicates viruses isolated in 2007.
Space, time, and host species were found to be significant predictors of the genetic distances (ORs > 1.0) for each of the gene segments without any two-way interactions, as indicated by the lowest deviance information criterion and the significance of the probability interval for the ORs (Table 2). The values of the OR in this study provide the relative change of the odds [(OR – 1) * 100] under two different condition of a single predictor. For example, time was a significant predictor of the genetic distances of the HA gene, in which an OR = 1.37 indicate a 37% increase of the odds of all genetic distances of the HA gene of two pairs of isolates being different, when the difference in their sampling time increases by 1 standardized unit of time (Table 2). Model validation for all of three gene segments suggested a good fit as indicated by the results of R, R*, and paired t-test (Table 3). No outliers were detected in the series of paired predicted and observed values for all of the three gene segments in the validation subset (Grubb test, P < 0.05).
Table 2.
Association between space, time, and host species of H5N1 HPAIV outbreaks and the pairwise number of different nucleotides in the HA, NA, and NS genes of 70 isolates obtained from birds during multiple outbreaks between 2005 and 2007 in Romania.A
| Probability interval (%) |
|||
|---|---|---|---|
| Epidemiological factor | ORB | 2.5 | 97.5 |
| HA gene | |||
| Distance | 1.04 | 1.01 | 1.08 |
| Time | 1.37 | 1.32 | 1.43 |
| Species | 1.22 | 1.15 | 1.30 |
| NA gene | |||
| Distance | 1.1 | 1.07 | 1.14 |
| Time | 1.11 | 1.07 | 1.15 |
| Species | 1.11 | 1.04 | 1.17 |
| NS gene | |||
| Distance | 1.15 | 1.11 | 1.20 |
| Time | 1.25 | 1.20 | 1.31 |
| Species | 1.09 | 1.02 | 1.17 |
Distance and time standardized (subtracting the mean and dividing by the standard deviation).
Odds ratios (OR) represents an increase in the odds of all nucleotide of HA, NA, or NS genes of two isolates being different when the difference in the value of the predictor increases by 1 standardized unit.
Table 3.
Model validation results for the association between space, time, and host species of H5N1 HPAIV outbreaks and the number of different nucleotides in the HA, NA, and NS genes of 70 isolates sampled from birds during multiple outbreaks between 2005 and 2007 in Romania.A
| Gene segment | Mean OD% (95% CI) | Mean PD% (95% CI) | Paired t-test P-value | R | R P-value | R* |
|---|---|---|---|---|---|---|
| HA | 0.96 (0.93, 0.99) | 0.91 (0.88, 0.95) | 0.99 | 0.65 | <0.05 | 0.010 |
| NA | 0.66 (0.64, 0.69) | 0.55 (0.55, 0.56) | 1.00 | 0.66 | <0.05 | 0.013 |
| NS | 0.69 (0.66, 0.73) | 0.58 (0.56, 0.59) | 1.00 | 0.75 | <0.05 | 0.004 |
PD = predicted genetic distances, OD = observed genetic distances, R = Spearman correlation, R* = absolute value of the shrinkage on cross-validation, CI = confidence interval.
DISCUSSION
Results of this study suggest that an increase in the variation between any pair of H5N1 HPAIVs, as measured by the number of nucleotide differences between HA, NA, and NS genes, was associated with the geographical locations, periods of time, and host species of the outbreaks. In each of the three gene segments of the 70 H5N1 HPAIV, a high probability of nucleotide differences (ORs > 1) was found between those viruses sampled from places at greater distances from each other, viruses sampled over greater periods of time, and viruses derived from different species (Table 2). The results of the present study agree with previous ecological and genomic studies that described how H5N1 HPAIV is under continuous evolution through rapid nucleotide substitution rates of its HA, NA, and NS genes, at different geographical locations, periods of time, and host species (2,7,10,11,15,20,30,31). Furthermore, the strength of the association (ORs) between the model’;s predictors and the chances of nucleotide substitutions was of similar magnitude for the HA and NS genes (Table 2), suggesting that the nucleotide substitution rate was consistent and occurred simultaneously in two or more gene segments of H5N1 HPAIV sampled from different outbreaks in Romania between 2005 and 2007. Those results support findings from earlier studies that suggest that there may be equal magnitudes in the probability of nucleotide substitution occurring simultaneously in different gene segments of any given H5N1 HPAI isolate (31,33).
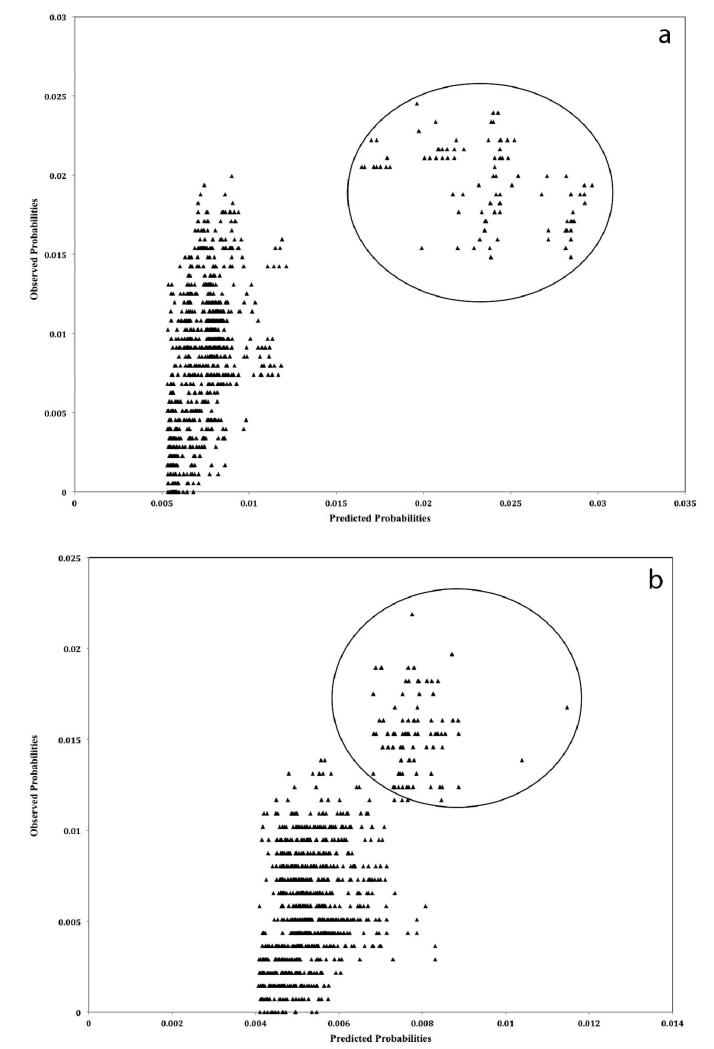
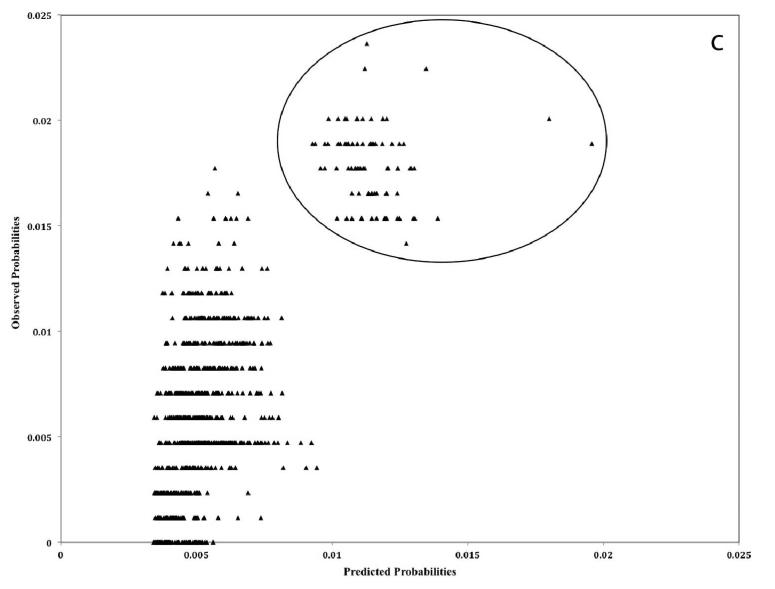
Phylogenetic analysis and correlations between the OD and PD suggested that the 70 H5N1 HPAIV obtained from birds across Romania between 2005 and 2006 were closely related (Figs. 3, 4). However, the H5N1 HPAIV from 2007 was less closely related to the 2005 and 2006 viruses (Figs. 3, 4). These findings agree with the notion that the Romanian outbreaks may have resulted from further multiple introductions during 2005–2007, as described elsewhere (9). The models of each gene segment have shown better prediction for the genetic distances of the 2007 H5N1 HPAIVs than those of the 2006 and 2005 viruses (Fig. 4), which could be attributed to the very small genetic differences of the 2006 and 2005 H5N1 HPAIVs. In this study, four out of the eight bird species were represented by only one isolate, and three others were represented by <8 isolates (Table 1). For that reason, the association between individual bird species and nucleotide variation of HPAIV was not assessed. Instead, the association between pairwise difference in the species of isolation and nucleotide substitution was assessed, and each pair of samples was categorized as either isolated from the same or different bird species.
Fig. 4.
Observed and predicted genetic distances (a = HA gene, b = NA gene, c = NS gene), expressed as a percentage, between 70 H5N1 HPAIV isolates sampled from birds during multiple outbreaks between 2005 and 2007 in Romania. The circle indicates the pairs corresponding to the 2007 isolates.
The application of the modeling approach here for predicting genetic distances of virus isolates obtained from an AIV outbreak may depend on the quality of the sequencing data. For example, in the present study, few of the HA gene sequences were missing some nucleotide information which has resulted in unequal alignment. Furthermore, the median number of different nucleotides between the 2005–2006 isolates was very small in each of the three genes. In the model here it was assumed that nucleotide substitutions occurred independently from those in other genes, which ignores the observation that the nucleotide substitution process is dependent between all genes (11,16,26). Although an extension of the current model to account for such dependence is straightforward, in the course of an H5N1 HPAIV epidemic, human and financial resources for full genome sequences may not be available. Therefore, a model predicting genetic distances for one gene segment such as the one presented here is likely to have broader application than a full genome model. Utility of such models in predicting genetic distance may also depend on the quality and quantity of epidemiological data obtained in the field during the course of an epidemic. For example, in the present study, the effect of poultry density, wild bird abundance, and weather conditions during the outbreaks, which are believed to be important epidemiological factors (2,7,11), could not be assessed because such data were not available.
The modeling approach presented may be useful, for example, during the course of an outbreak to predict genetic distances from the index virus to secondary outbreaks, by only using epidemiological data obtained in the field, and in the absence of genetic information from the secondary cases. Thus, without the need for detailed sequence data for the secondary outbreaks, some inferences may be made about the evolution of the virus and how closely related new outbreak isolates are to previously collected isolates; when compared to the actual sequence from secondary outbreaks, inferences on the probability of new incursions may be made (22). Therefore, the presented model could complement traditional molecular characterization methods that commonly account for only temporal factors by providing further insights into host and environmental risk factors associated with the variation in the number of different nucleotides of prevailing epidemic strains of H5N1 HPAIV. However, in the face of multiple outbreaks, where a lack of resources often precludes rapid nucleotide sequence analysis of isolates, the model could be used to identify which outbreaks might not be caused by closely related viruses and to help establish priorities for sampling and nucleotide analysis.
In conclusion, the model proposed in the present study could be useful in estimating and understanding the degree of relatedness between strains, and could provide additional information to that available from traditional phylogenetic trees in the face of an AIV epidemic. Furthermore, our model uses epidemiological and genetic data not only to predict genetic distances between H5N1 HPAIV, but also to measure deviations from expected genetic differences; thus our approach may also predict new virus introductions or emergence in the face of an outbreak.
ACKNOWLEDGMENTS
The authors thank Dr. Laural Beckett and Dr. Xiaowei Yang (Department of Public Health Sciences, University of California, Davis, CA) for revisions and feedback on the study. This work was supported by the EU Reference Laboratory for Avian Influenza and the Wellcome Trust, London, United Kingdom.
Abbreviations
- AIV
avian influenza virus
- dni,j
number of different nucleotides between each pair of isolates
- EU
European Union
- f
independent fixed effect
- HPAIV
highly pathogenic avian influenza virus
- KMi,j
pairwise spatial distance
- N
number of all possible pairs of the isolates
- OD
observed genetic distance
- OR
odds ratio
- p
probability of nucleotide differences
- PD
predicted genetic distance
- R
Spearman correlation coefficient
- R2
sample squared multiple correlation
- R*
absolute value of the shrinkage on cross validation
- Spi,j
pairwise difference in the species of isolation
- Ti,j
pairwise absolute difference in time
- U
nonstructured random effect
- WinBUGS
Bayesian inference by using Gibbs sampling
- α
model intercept
- β
regression coefficients for each predictor
References
- 1.Air GM, Laver WG. The neuraminidase of influenza virus. Proteins. 1989;6:341–356. doi: 10.1002/prot.340060402. [DOI] [PubMed] [Google Scholar]
- 2.Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine. 2007;25:5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 3.Breed AC, K Harris,, Hesterberg U, Gould G, Londt BZ, Brown IH, Cook AJ. Surveillance for avian influenza in wild birds in the European Union in 2007. Avian Dis. 2010;54:399–404. doi: 10.1637/8950-053109-Reg.1. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Li Y, Li Z, Shi J, Shinya K, Deng G, Qi Q, Tian G, Fan S, Zhao H, Sun Y, Kawaoka Y. Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J. Virol. 2006;80:5976–5983. doi: 10.1128/JVI.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Deng YM. Influenza virus antigenic variation, host antibody production and new approach to control epidemics. Virol. J. 2009;6:30. doi: 10.1186/1743-422X-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grubbs FE. Procedures for detecting outlying observations in samples. Technometrics. 1969;11:1–21. [Google Scholar]
- 7.Guan Y, Smith GJ, Webby R, Webster RG. Molecular epidemiology of H5N1 avian influenza. Rev. Sci. Tech. 2009;28:39–47. doi: 10.20506/rst.28.1.1868. [DOI] [PubMed] [Google Scholar]
- 8.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program. Version 7.0.9. Nucl. Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 9.Howard W, Watson S, Baillie G, Alkhamis M, Franz S, Focosi-Snyman R, Perez A, Iuliana O, Kellam P, Brown I. Evolutionary dynamics and molecular characterisation of H5N1 highly pathogenic avian influenza viruses from Romania 2005-2010; 9th International Congress of Veterinary Virology. European Society for Veterinary Virology; Madrid. 2012.p. 174. [Google Scholar]
- 10.Ibrahim MS, Watanabe Y, Ellakany HF, Yamagishi A, Sapsutthipas S, Toyoda T, Abd El-Hamied HS, Ikuta K. Host-specific genetic variation of highly pathogenic avian influenza viruses (H5N1) Virus Genes. 2011;42:363–368. doi: 10.1007/s11262-011-0583-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klenk HD, Garten W, Matrosovich M. Molecular mechanisms of interspecies transmission and pathogenicity of influenza viruses: lessons from the 2009 pandemic. BioEssays. 2011;33:180–188. doi: 10.1002/bies.201000118. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li KS, Guan Y, Wang J, Smith GJ, Xu KM, Duan L, Rahardjo AP, Puthavathana P, Buranathai C, Nguyen TD, Estoepangestie AT, Chaisingh A, Auewarakul P, Long HT, Hanh NT, Webby RJ, Poon LL, Chen H, Shortridge KF, Yuen KY, Webster RG, Peiris JS. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 14.Lunn DJ, Thomas A, Best N. Statistics and computing, 1.4.3 ed. Imperial College and Medical Research Council; London, United Kingdom: 2000. WinBUGS—a Bayesian modelling framework; concepts, structure, and extensibility. [Google Scholar]
- 15.Lvov DK, Shchelkanov MY, Prilipov AG, Vlasov NA, Fedyakina IT, Deryabin PG, Alkhovsky SV, Grebennikova TV, Zaberezhny AD, Suarez DL. Evolution of highly pathogenic avian influenza H5N1 virus in natural ecosystems of northern Eurasia (2005–08) Avian Dis. 2010;54:483–495. doi: 10.1637/8893-042509-Review.1. [DOI] [PubMed] [Google Scholar]
- 16.Medina RA, Garcia-Sastre A. Influenza A viruses: new research developments. Nat. Rev. Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhtar MM, Rasool ST, Song D, Zhu C, Hao Q, Zhu Y, Wu J. Origin of highly pathogenic H5N1 avian influenza virus in China and genetic characterization of donor and recipient viruses. J. Gen. Virol. 2007;88:3094–3099. doi: 10.1099/vir.0.83129-0. [DOI] [PubMed] [Google Scholar]
- 18.Nakajima K, Nobusawa E, Nakajima S. Evolution of the NS genes of the influenza A viruses. II. Characteristics of the amino acid changes in the NS1 proteins of the influenza A viruses. Virus Genes. 1990;4:15–26. doi: 10.1007/BF00308562. [DOI] [PubMed] [Google Scholar]
- 19.Nakajima K, Nobusawa E, Ogawa T, Nakajima S. Evolution of the NS genes of the influenza A viruses. I. The genetic relatedness of the NS genes of animal influenza viruses. Virus Genes. 1990;4:5–13. doi: 10.1007/BF00308561. [DOI] [PubMed] [Google Scholar]
- 20.Neumann G, Chen H, Gao GF, Shu Y, Kawaoka Y. H5N1 influenza viruses: outbreaks and biological properties. Cell Res. 2010;20:51–61. doi: 10.1038/cr.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oprisan G, Coste H, Lupulescu E, Oprisoreanu AM, Szmal C, Onita I, Popovici N, Ionescu LE, Bicheru S, Enache N, Ceianu C, Czobor F, Olaru E, Alexandrescu V, Radu DL, Onu A, Popa MI. Molecular analysis of the first avian influenza H5N1 isolates from fowl in Romania. Roum. Arch. Microbiol. Immunol. 2006;65:79–82. [PubMed] [Google Scholar]
- 22.Perez AM, Konig G, Spath E, Thurmond MC. Variation in the VP1 gene of foot-and-mouth disease virus serotype A associated with epidemiological characteristics of outbreaks in the 2001 epizootic in Argentina. J. Vet. Diagn. Invest. 2008;20:433–439. doi: 10.1177/104063870802000404. [DOI] [PubMed] [Google Scholar]
- 23.Perez AM, Pauszek SJ, Jimenez D, Kelley WN, Whedbee Z, Rodriguez LL. Spatial and phylogenetic analysis of vesicular stomatitis virus over-wintering in the United States. Prev. Vet. Med. 2010;93:258–264. doi: 10.1016/j.prevetmed.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Perez AM, Zeng D, Tseng CJ, Chen H, Whedbee Z, Paton D, Thurmond MC. A Web-based system for near real-time surveillance and space-time cluster analysis of foot-and-mouth disease and other animal diseases. Prev. Vet. Med. 2009;91:39–45. doi: 10.1016/j.prevetmed.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Proenca-Modena JL, Macedo IS, Arruda E. H5N1 avian influenza virus: an overview. Braz. J. Infect. Dis. 2007;11:125–133. doi: 10.1590/s1413-86702007000100027. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki Y. Avian and human influenza virus receptors and their distribution. Adv. Exp. Med. Biol. 2011;705:443–452. doi: 10.1007/978-1-4419-7877-6_23. [DOI] [PubMed] [Google Scholar]
- 27.Takeda M, Leser GP, Russell CJ, Lamb RA. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for mulitple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandegrift KJ, Sokolow SH, Daszak P, Kilpatrick AM. Ecology of avian influenza viruses in a changing world. Ann. N. Y. Acad. Sci. 2010;1195:113–128. doi: 10.1111/j.1749-6632.2010.05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijaykrishna D, Bahl J, Riley S, Duan L, Zhang JX, Chen H, Peiris JS, Smith GJ, Guan Y. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathogens. 2008;4:e1000161. doi: 10.1371/journal.ppat.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe C, Uchida Y, Ito H, Ito T, Saito T. Host immune-related gene responses against highly pathogenic avian influenza virus infection in vitro differ among chicken cell lines established from different organs. Vet. Immunol. Immunopathol. 2011;144:187–199. doi: 10.1016/j.vetimm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Williams RA, Peterson AT. Ecology and geography of avian influenza (HPAI H5N1) transmission in the Middle East and northeastern Africa. Int. J. Health Geog. 2009;8:47. doi: 10.1186/1476-072X-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization for Animal Health (OIE) [cited Nov 2012];World Animal Health Information Database (WAHID) Interface, weekly disease information. 2012 Available from: http://www.oie.int/wahis_2/public/wahid.php/Wahidhome/Home.
- 35.World Health Organization/Office International des Epizooties/Food and Agricultural Organization H5N1 Evolution Working Group Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1) Emerg. Infect. Dis. 2008;14:e1. doi: 10.3201/eid1407.071681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, Subbarao., Cox NJ, Guo Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology. 1999;261:15–19. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- 37.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza a viruses. J. Virol. 2009;83:10309–10313. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]