Figure 1.
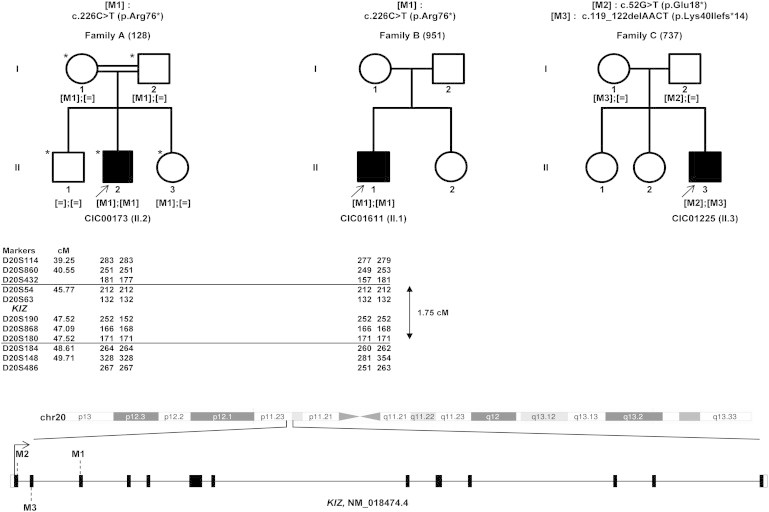
Identification of Homozygous and Compound-Heterozygous KIZ Variants in Three Unrelated Families Affected by Autosomal-Recessive RCD
Family A (left), family B (middle), and family C (right) were analyzed in the present study. Individuals highlighted with an asterisk were screened with WES. Affected individuals are marked with an arrow. A homozygous nonsense mutation, c.226C>T (p.Arg76∗), in KIZ was identified in the affected index subject from family A and cosegregated with the phenotype. Subsequent direct Sanger sequencing of KIZ in 340 individuals with autosomal-recessive RCD identified two other subjects with KIZ mutations: the affected index subject in family B had the homozygous c.226C>T (p.Arg76∗) mutation, and the affected index subject in family C had compound-heterozygous mutations c.52G>T (p.Glu18∗) and c.119_122delAACT (p.Lys40Ilefs∗14). Haplotype analysis performed for investigating whether the c.226C>T (p.Arg76∗) variant represents a founder mutation in families A and B is shown under each pedigree symbol. Microsatellite marker locations according to Marshfield genetic maps (Marshfield Laboratories) are shown in cM. Numbers indicate the allele size in nucleotides for each microsatellite. Both subjects were found to share a common haplotype of five polymorphic microsatellites (i.e., DS20S54, DS20S63, DS20S190, DS20S868, and DS20S180; delineated by two horizontal lines) flanking KIZ and spanning ≈1.75 cM (1.18 Mb). Filled and unfilled symbols indicate affected and unaffected status, respectively. Square boxes indicate males, and circles indicate females. In the schematic representation of the structure of KIZ (RefSeq NM_018474.4; harboring 13 exons), filled and unfilled boxes represent coding and noncoding exonic regions, respectively. M1, M2, and M3 depict the positions of the mutations identified in the current study.
