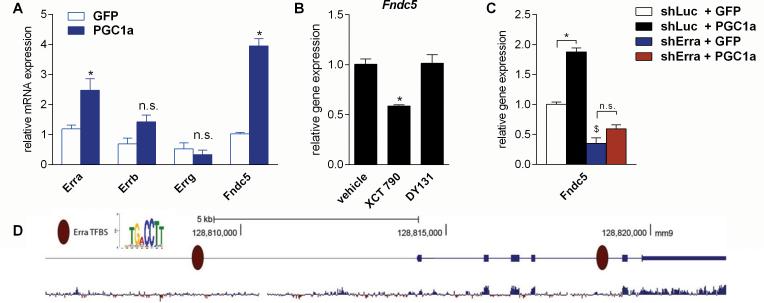
Figure 4. ERRα is a key interacting transcription factor with PGC-1α for regulating Fndc5 gene expression in neurons.
(A) Primary cortical neurons at DIV 7 were transduced with either PGC-1α or GFP adenovirus and harvested 48 hrs later. *P < 0.05 compared to control group.
(B) Primary cortical neurons at DIV 7 were treated with either XCT 790 (1 μM), a selective inverse ERRα agonist, DY131 (1 μM), a selective ERRβ and ERRγ agonist, or vehicle for overnight. *P < 0.05 compared to vehicle only group.
(C) Primary cortical neurons at DIV 4 were transduced with lentivirus carrying shRNA hairpins against either Erra or luciferase (Luc) as control. Three days later were cells were transduced with either PGC-1α or GFP adenovirus and harvested 48 hrs later. mRNA was prepared and gene expression was assessed by qPCR. Data are shown as mRNA levels relative to Rsp18 expression, expressed as mean ± SEM. *P < 0.05 compared to corresponding shLuc expressing control group. $P < 0.05 compared to corresponding GFP expressing control group. Data (A-C) are shown as mRNA levels relative to Rsp18 expression, expressed as mean ± SEM.
(D) Analysis of the murine Fndc5 promoter for putative ERREs. The murine Fndc5 gene and 6 kb of its upstream promoter were searched for the canonical ERRE: TGA CCTT. Genomic coordinates are given according to the assembly mm9 from the UCSC Genome Browser. The bottom diagram indicates the degree of mammalian conservation across the genomic locus. The presented motif was modified from www.factorbook.org (Wang et al., 2012). See also Figure S3.