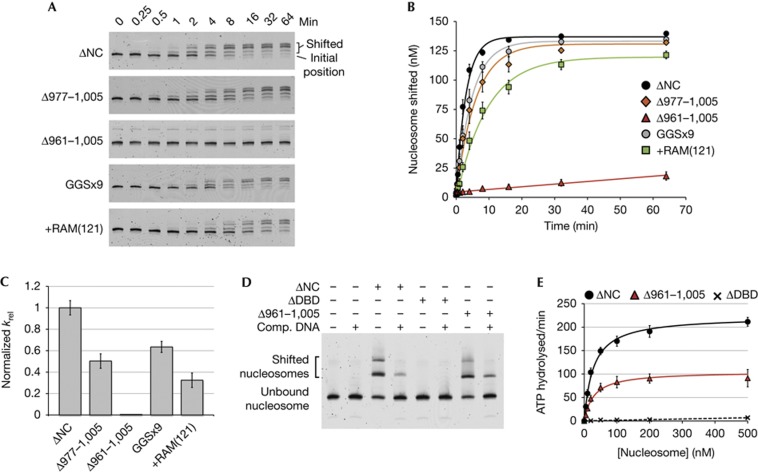
Figure 2.
Chd1 tolerates increased length and flexibility in the DBD linker, but requires a minimal length linker for coordinated action of DBD and ATPase domains. (A) Nucleosome sliding as assessed by native PAGE. Data shown are representative experiments (n≥4). (B) Quantification of nucleosome sliding reactions shown in (A). Error bars represent standard deviations (s.d.) (n≥4). (C) Relative rate constants for nucleosome sliding. Rate constants were obtained from single exponential fits to the data shown in (B), with the relative rates shown as krel=kobs, Chd1 variant/kobs, Chd1ΔNC. For Chd1ΔNC, kobs was 0.33±0.02 min−1. Error bars represent the calculated errors of the fits. (D) Deletion of the DBD linker does not disrupt nucleosome binding, as shown by native PAGE. Super-shifted bands indicate Chd1–nucleosome complexes. (E) Deletion of the DBD linker still allows the Chd1 ATPase motor to be stimulated by nucleosomes. Shown are the averages and s.d. (n≥3) of nucleosome-stimulated ATPase activities with increasing concentrations of nucleosome substrate. Nucleosome-free rates were subtracted from all values, and the resultant activities were fit to the Michaelis–Menton equation (Chd1ΔNC and Chd1ΔNCΔ961–1,005, solid lines) or linearly (Chd1ΔDBD, broken line). Chd1ΔDBD contained residues 118–1,005. DBD, DNA-binding domain; PAGE, polyacrylamide gel electrophoresis.