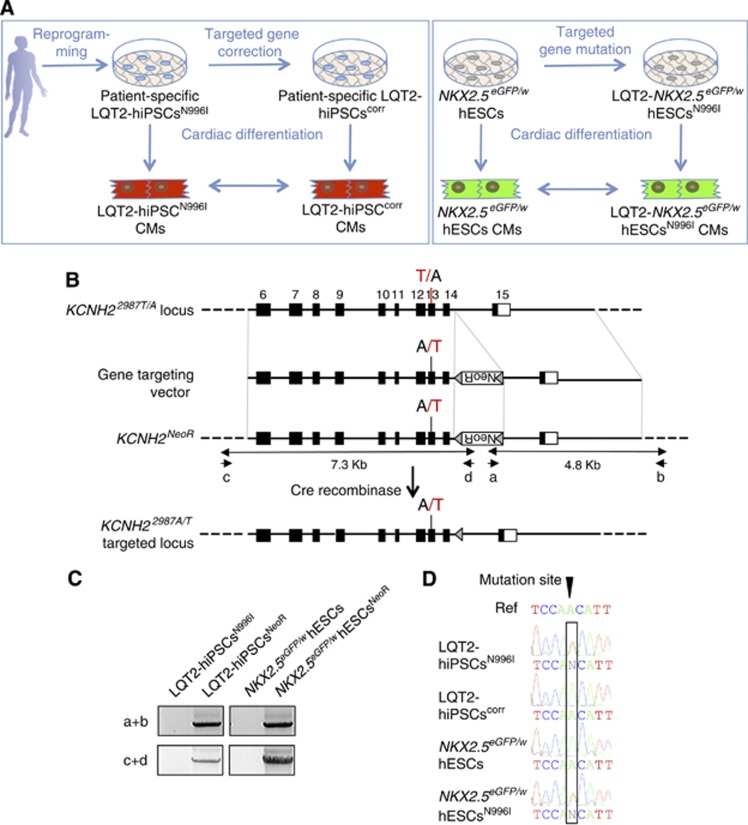
Figure 2.
Gene targeting by homologous recombination in LQT2-hiPSCsN996I and in NKX2.5eGFP/w hESCs. (A) Schematic showing the project rationale. Patient-specific LQT2-hiPSCs harbouring the N996I mutation were corrected and NKX2.5eGFP/w hESCs were mutated by gene targeting. Parental and genetically modified hPSC lines were differentiated into CMs and their electrophysiological phenotypes were analysed and compared. (B) The strategy for precise genomic modification of KCNH2. Top line, structure of the KCNH2 locus. Numbered black boxes indicate exons 6–15. Exon 13 is mutated (red T) in LQT2-hiPSCsN996I and wild type (black A) in NKX2.5eGFP/w hESCs. The gene targeting vector for correcting the mutation in LQT2-hiPSCs has the wild-type adenine nucleotide, whereas the gene targeting vector for introducing the mutation in NKX2.5eGFP/w hESCs has the mutated thymine nucleotide. NeoR, the PGK-Neo cassette encoding G418 resistance flanked by loxP sequences (grey triangles), was inserted in the reverse direction. PCR primers (a, b) and (c, d) were used to identify the targeted clone. (C) PCR analysis using these primers generated specific bands of 4.8 kb (5′ homology arm) and 7.3 kb (3′ homology arm) from targeted clones (LQT2-hiPSCsNeoR and NKX2.5eGFP/w hESCsNeoR). (D) Sequence analysis of PCR-amplified genomic DNA showing correction of the c.A2987T mutation in the LQT2-hiPSCscorr line and mutation in the NKX2.5eGFP/w hESCsN996I line. The wild-type reference sequence (Ref) is shown in the top line.
Source data for this figure is available on the online supplementary information page.