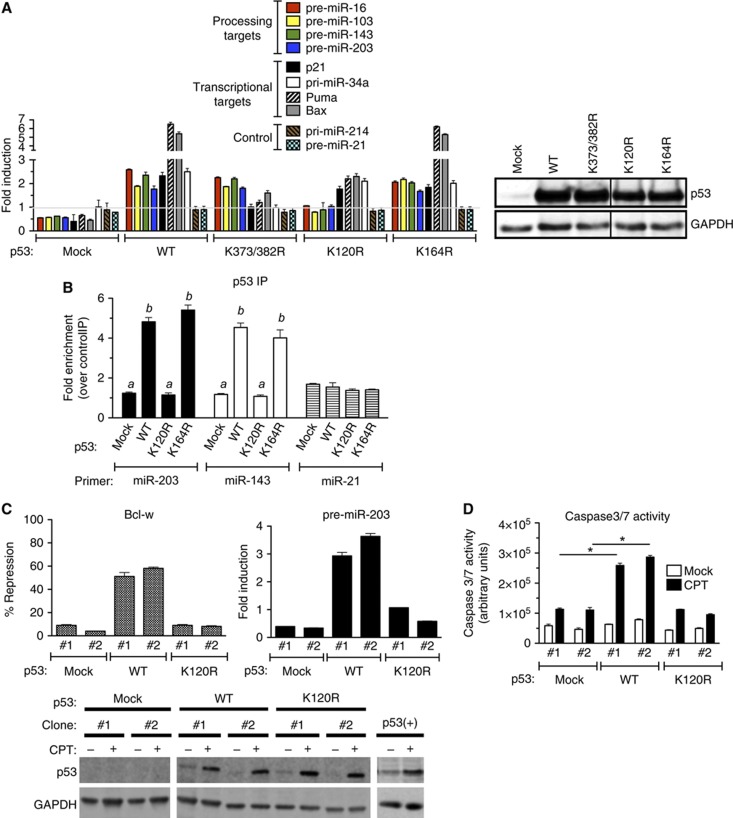
Figure 4.
Modification at the K120 residue is critical for the miRNA processing activity of p53. (A) p53(−) cells were transfected with empty expression vector (mock), expression vector carrying wild-type p53 (WT), K120R mutant, K164R mutant, or K373/382 double mutant (K373/382R), followed by DMSO or CPT treatment for 16 h. Total RNA was subjected to qRT–PCR analysis of p53-processed targets (miR-16, 103, -143, and -203), p53 transcriptional target (p21, Puma, Bax, and pri-miR-34a), or control (pri-miR-214 and pre-miR-21). Results are shown as the fold change of pre-miRNA upon CPT treatment as compared to DMSO treatment (left panel). Total lysates of the same cells or p53(+) cells following DMSO treatment were subjected to immunoblot analysis of p53 and GAPDH (loading control; right panel). (B) RNA-IP was performed on p53(−) cells transfected with the expression vector carrying p53 WT, K120R, or K164R. pri-miRNA levels of processing targets of p53 (miR-203 and miR-143) or control (miR-21) associated with p53 WT or mutants were analysed by qRT-PCR. Results are shown as the fold induction of p53-associating pri-miRNAs upon CPT treatment for 8 h over DMSO treatment. Immunoblot analysis of p53 and GAPDH is shown in (B). (C) Two independent clones of p53(−) cells stably expressing an empty vector (mock), WT, or K120R were treated with DMSO (mock) or CPT for 16 h and subjected to qRT-PCR analysis of pre-miR-203 and Bcl-w mRNA normalized to GAPDH. Result is shown as % repression and fold induction of Bcl-w and pre-miR-203 by comparing the levels in CPT-treated cells with DMSO-treated cells. The levels of p53 and GAPDH (loading control) protein following DMSO (mock) or CPT treatment were examined by immunoblotting using the same stable clones. (D) p53(−) cells stably expressing empty vector (mock), WT, or K120R were treated with DMSO (mock) or CPT for 16 h and subjected to the caspase-3/7 activity assay. *P<0.01; error bars represent standard deviation (s.d.). n=3.
Source data for this figure is available on the online supplementary information page.