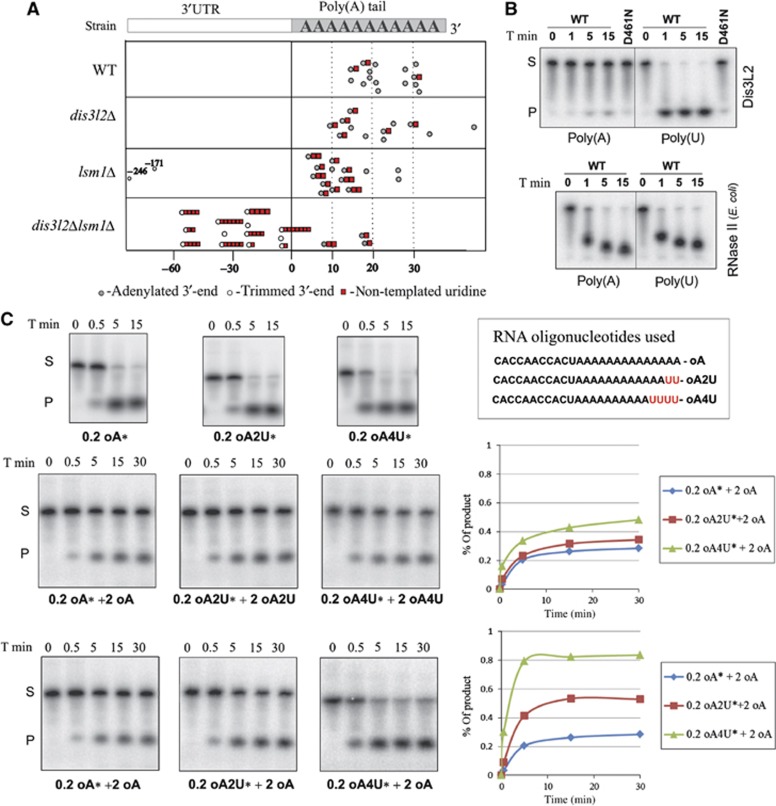
Figure 7.
Dis3L2 preferentially degrades uridylated RNAs. (A) Deletion of dis3l2+ in Δlsm1 background results in the accumulation of trimmed and uridylated adh1+ transcripts. Each circle point in the table indicates the position of one adh1+ mRNA 3′-end. Point zero represents the standard polyadenylation site of adh1+ mRNA. Positive values correspond to the length of poly(A) tail attached to the polyadenylation site, and the negative values correspond to the 3′-end position in 3′-UTR if mRNA was trimmed. Uridylation is indicated by the red squares, and number of squares corresponds to the number of non-templated U residues detected at the 3′-end. (B) Dis3L2 exhibits stronger preference for poly(U) compared to poly(A) RNA substrate in vitro. Dis3L2 protein (WT) and mutated version (D461N) were incubated with the same amounts of radioactively labelled poly(A) or poly(U) RNAs. Reaction was stopped at the indicated time points and products were separated on denaturing polyacrylamide gels. The same substrates were incubated with bacterial RNase II. (C) Uridine residues added to the 3′-end can effectively target RNA substrates for degradation by Dis3L2 in vitro. Same amounts of Dis3L2 protein were incubated with the indicated amounts of different RNA substrates (0.2 or 2 pmol). Substrate sequence is indicated. Reactions with radioactive substrates (labelled with *) were supplemented with the substrates that were not radioactively labelled. Reactions were stopped at the indicated time points (T min) and products were separated on denaturing polyacrylamide gels. The graphs (on the right side) depict the accumulation of the product at different time points as calculated using Image Quant.
Source data for this figure is available on the online supplementary information page.